Prof. Dr. M. Bartkuhn
 |
Prof. Dr. Marek Bartkuhn Biomedical Informatics and Systems Medicine Science Unit for Basic and Clinical Medicine Aulweg 128, Room 111 |
|---|
Applied bioinformatics approaches for epigenomics.
We study DNA-templated nuclear processes from a genome-wide perspective. Amongst these processes our main interest is how gene expression is regulated and how chromatin helps, modulates or restricts gene activity.
Transcription of our genes happens in the context of chromatin, a nucleo-protein complex, into which DNA is packaged inside the nuclei of our cells. The basic building block of chromatin is the nucleosome, consisting of an octamer of histones, around which the DNA is wrapped in 1 3/4 turns, corresponding to around 146 bp of DNA. By this, DNA can be compacted and further compaction is achieved by folding the nucleosomal building blocks into higher order structures. As a consequence, the 2 meters of DNA fit into a 10 µm cell nucleus. On the other hand, this packaging makes DNA highly inaccessible for processes like transcription, repair and replication. A complex machinery of transcription factors, chromatin remodeling, DNA- and histone-modifying enzymes has emerged during evolution in order to regulate accessibility of chromatin. This layer of genome regulation is summarized in the term epigenome.
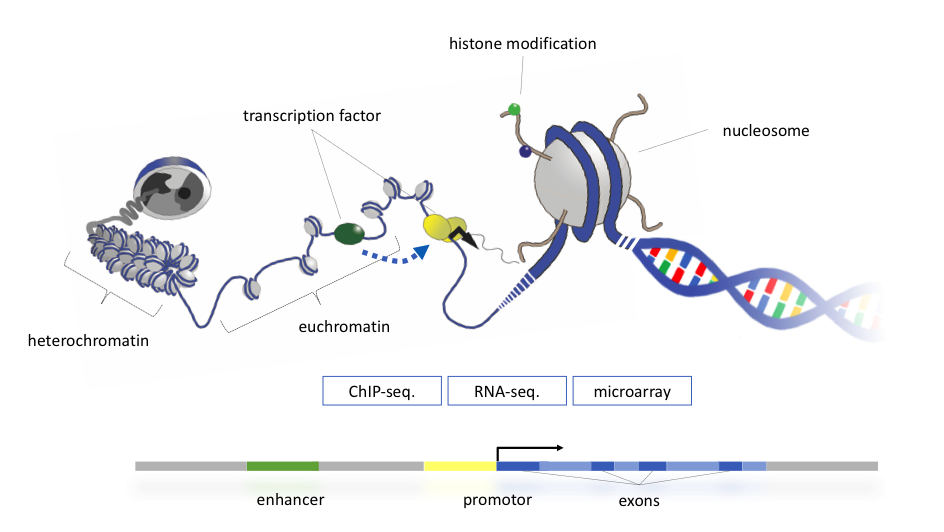
Figure 1: Schematic overview about the multi-dimensional relationships relevant for the for the transcriptional output of a gene. Nowadays, many of the parameters can be measured on genome-wide scale.
Here, bioinformatics come into play for two reasons: first, modern experimental approaches involve the generation of very large data sets. Processing of such data sets requires specific computing infrastructures with respect to hardware as well as software requirements. Second, the interpretation of the data becomes more and more challenging, majorly due to the fact, that most of the modern approaches deliver data on genome-wide scale. In case of gene regulation the term „genome-wide“ indicates, that instead of studying the regulation of individual model genes in order to identify general regulatory principles, we rather study the complete set of genes that exist in our genomes at the same time. Current methods involve the application of next generation sequencing methods in order to derive estimates for the expression levels of all the genes in our genome in one go (RNA-seq). Likewise, we can measure many other parameters of chromatin on genome-wide scale, such as where and how much of a given transcription factor or other chromatin associated regulatory proteins bind to specific genomic regions, where specific chromatin modifications are located (ChIP-seq) or how DNA accessibility is distributed across the genome. Additionally, methods based on chromosome conformation capture based techniques (3C) allow us to study the overall 3-dimensional-conformation of our genome (Hi-C). We try to integrate data from different sources in order to better understand the complex network of epigenetic regulatory processes.

Figure 2: Portfolio of different genome-wide data sets that were recently analyzed by our group – often in integrative fashion.
Although regulation of transcription is clearly our main focus, we are basically interested in many different aspects of genome biology. This includes replication and repair and may also include other processes like RNA-processing, which is known to depend on chromatin, too.
Usually we apply our approaches within collaborations together with experimental biologists. Within these collaborations we
1) help collaborators with experimental and statistical design of their studies
2) analyze genome-wide data sets
3) integrate different sources of data in systems biology approaches
4) represent analyzed data in form of state of the art figures for publication
5) develop, improve and provide automated pipelines in order to make analysis tools and workflows available to collaborators (via a Galaxy server)
Additionally, we are interested in improving existing methods or in case not available, the development of new analysis strategies and tools.
We are always looking for motivated bachelor or master candidates. Please apply irrespective of previous experience with bioinformatics approaches. Basic knowledge of programming is a plus but not a must.
