AG Hake
Chromatin: Packaging and regulatory structure of the eukaryotic genome
All eukaryotes package their genome in form of chromatin. The smallest unit of chromatin is the nucleosome that consists of DNA wrapped around an octamer of histone proteins. Besides “just” being packaging material, most importantly, chromatin has a functional role as its structure and composition takes part in regulating gene expression, repair of DNA damage, cell cycle progression, apoptosis, genome maintenance and stability. Thereby, chromatin shapes the epigenetic feature of a cell and influences cell differentiation and organism development.
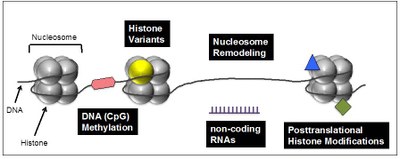
Several interconnected mechanisms have evolved that influence chromatin structure in a well-coordinated manner. Besides DNA methylation, chromatin remodeling, noncoding RNAs and posttranslational histone modifications, the exchange of canonical histones with their specialized variant counterparts are important means to regulate chromatin in general and access of e.g. transcription factors to the underlaying DNA sequence in particular (Figure 1).
|
Figure 1: Chromatin-modifying processes involved in regulation of DNA-based processes. Chromatin, consisting of nucleosomes (histone octamer + 145 bp DNA), is not only DNA packaging material in eukaryotes but does also play an important in role in regulating transcription, DNA damage repair and genome integrity. Changes in DNA accessibility though difference in chromatin structure are achieved through interconnected and reversible processes including DNA methylation (red hexagon), exchange of canonical histones (gray sphere) with specialized histone variants (yellow sphere), nucleosome remodeling and sliding, non-coding regulatory RNAs (purple) and posttranslational histone modifications (blue triangle and green square). |
|---|
Core histone variants: Modulators of chromatin structure and function
We are interested in understanding the functional role of human histone variants and their deposition machineries (chaperone and/or remodeling complexes). Histone variants are expressed in a replication-independent manner and affect chromatin and cellular processes by different means. Histone variants are deposited into chromatin via dedicated chaperone / remodeling complexes in a spatially and temporally restricted and tightly controlled manner.
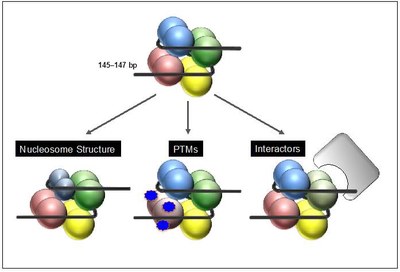
After their deposition into chromatin in exchange with canonical histones variants can either change nucleosome structure and DNA accessibility directly, or influence chromatin indirectly by altering posttranslational modification pattern and/or mediate the recruitment of variant-specific binding proteins that in turn modify the surrounding chromatin environment (Figure 2).
|
Figure 2: Function of histone variants. After chromatin deposition histone variants can affect biological processes by different means. Some variants directly affect nucleosome structure leading to a gain or loss of DNA accessibility due to changes in DNA binding (left). In addition, histone variants can indirectly influence DNA-based processes by changes of their posttranslational modification (PTM) pattern and/or by recruiting variant-specific interactors that in turn might modify the surrounding chromatin environment. |
|---|
Goals
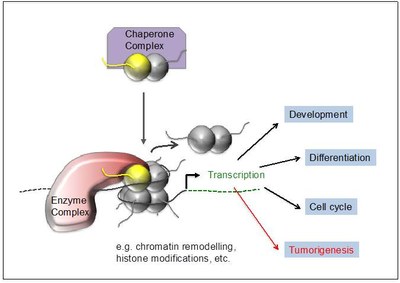
Our long-term goals are to understand how histone variants regulate DNA-based processes such as gene expression, DNA damage repair and cell cycle control and how deregulation and mutations of such mechanisms contribute to tumorigenesis (Figure 3).
|
Figure 3: Function of histone variants. Chaperone complexes deposit histone variants in exchange with their canonical counterpart into chromatin, possibly leading to the recruitment of variant-interacting chromatin complexes. They modify the surrounding chromatin structure affecting among others transcription which in turn regulates e.g. development, cell differentiation and cell cycle progression. Deregulation or mutation of the histone variant itself or components of its interacting network can contribute to or even drive tumorigenesis. |
|---|