Current topics of our group
As members of a pharmacology institute we teach the entire spectrum of drugs applied to humans including the complex mechanisms of action of all antiinflammatory drugs used today.
In contrast, our resarch is focussed on as yet poorly understood aspects of signal transduction of IL-1, TNF and various stressful stimuli. This includes the mechanisms of cytokine-mediated reprogramming of cellular gene expression which lead to activation of many additional inflammatory genes thus forming the basis of any cellular inflammatory reaction. These processes also play fundamental roles in bacterial or viral infections or stress reactions. Often, these triggers make redundant use of signaling molecules. Therefore, our investigations on the IL-1 system can easily be connected to the major reserach topics of our university such as the Excellence Cluster Cardiopulmonary System (ECCPS) or the Universities of Giessen Marburg Lung Center (UGMLC).
Control of gene expression requires a complex interplay of multiple signaling pathways during initiation and termination of mRNA synthesis in the nucleus and mRNA decay in the cytoplasm. Hence, we study both, the nuclear signaling processes occuring at chromatin as well as protein:RNA complexes within the cytoplasm. A major focus are also reversible posttranslational modifications on signaling proteins such as phosphorylations, ubiquitinations or acetylations which serve as fast molecular switches to activate or terminate protein functions in inflammatory reactions. These analyses require model systems providing sufficient numbers of cells. Thus, we are using human or murine transformed or immortalized cell lines. By the biochemical, molecular biology, cell biology and bioinformatics methods established in our lab, we can follow the entire flow of signals starting from the receptor down to the nucleus. In many cases we can perfom quantitative measurements. By using modern molecular biology techniques including various forms of RNAi and genome editing by the CRISPR-Cas9 system we manipulate individual steps of signal transduction trying to establish causal relationsships and to delineate mechanisms.
The followig scheme shows a simplified version of IL-1 signal transduction within a cell as we understand it today. The integrated links lead to results that we are obtaining in our lab at several levels of IL-1 signaling and demonstrate our options to study IL-1 signal transduction at high molecular resolution.
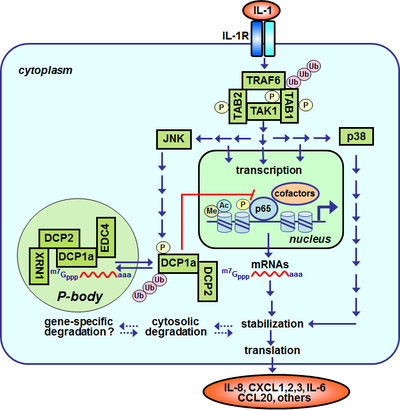
Schematic representation of aspects of IL-1 signal transduction that are studied in our lab
The heterodimeric IL-1 receptors (IL-1R) are constitutively expressed on most cells of our body. In contrast, the agonistic ligands (IL-1α and IL-1β) are newly synthezised or released by cells of the innate immune system (e.g. monocytes, macrophages) during inflamamtory reactions. Binding of IL-1α or IL-1β to the IL-1R induces through protein:protein interactions (not shown) and receptor proximal adaptor proteins and protein kinases (MYD88, IRAK1-4, Tollip) within 2-10 minutes the formation of oligomeric signaling complexes comprising the adaptor protein TRAF6, the protein kinase TAK1 and the TAK1-interacting proteins TAB1-TAB3. TARF6 functions as an ubiquitin E3 ligase catalyzing the synthesis ubiquitin chains which stabilize and promote protein:protein interactions with additional effector molecules of the signaling pathway (for an example see here). TAK1 and TAB1-TAB3 become reversibly phosphorylated in response to IL-1. An experiment demonstrating the rapid IL-1-dependent formation of this signaling complex is shown here (activation nof TAK1). After a delay of 10-30 Minuten the MAP kinases JNK, p38 and ERK (not shown) and the NF-κB signaling pathway are activated (activation of p65 (1), (2)). JNK phosphorylates and activates transcription factors of the AP-1 family such as c-Jun (activation of JNK). Cells lacking c-Jun or JNK display strong defects in IL-1-mediated gene regulation (see here). NF-κB is a transcription factor with complex regulation which in its inactive form is retained within the cytosol and which upon IL-1 treatment translocates to the nucleus where it binds to specific NF-κB sites in the regulatory regions (promoters, enhancers) of many inflammatory genes. For an example of IL-1-dependent binding of NF-κB to the IL8 promoter see here. For an overview on IL-8 gene regulation see here. For an example experiment addressing genome-wide changes of IL-1-induced gene expression see visualization of a microarray experiment. For an example of the genome-wide investigation of protein:DNA complexes by ChIP-seq see here. In addition to gene transcription, IL-1 induces the formation of cytoplasmic processing-bodies which consist of protein:RNA complexes and contain decapping and other mRNA decay factors (for an example see here). The known part of the IL-1 signaling system comprises around 100 components which we have combined to a network and connected with mathematical operators in order to better model the signaling flow. This signaling system crosstalks at multiple levels with other signaling networks such as the cell cycle (see IL-1, cell cycle and CDK6).
Abbreviations: IL-1R, interleukin-1 receptor; TRAF6, TNF-receptor-associated factor 6; TAK1, TGFβ-activated protein kinase 1; TAB1/2, TAK1-binding protein1/2; JNK, Jun-N-terminal kinase; NF-κB, nuclear factor kappa B; Pol II, RNA polymerase II; P, phosphorylation; ac, acetylation; ub, ubiquitination; XRN1, 5'-3' Exoribonuclease 1; DCP1a, decapping enzyme homolog 1 a; DCP2, decapping enzyme homolog 2; EDC4, enhancer of mRNA decapping 4.
