Research Details, Wende
- Highly specific tailor-made nucleases as essential tools for genome engineering
-
Highly specific tailor-made nucleases as essential tools for genome engineering
After the discovery of the DNA as the molecule carrying the hereditary information of all living organisms, DNA processing enzymes, in particular restriction endonucleases, led to the development of the recombinant DNA technology [1]. Today, in the post-genomic era, a steadily growing number of entire genome sequences is available. The emerging genome editing and genome reprogramming technologies require more sophisticated tools with very high specificity, ideally targeting a unique sequence in a complex genome [2]. Both technologies are highly innovative and have numerous applications in basic science, biotechnology and medicine. These days, transcription activator like effector (TALE) nucleases and the recently introduced CRISPR-Cas9 system have widely spread and become the methods of choice for precision genome engineering [3].
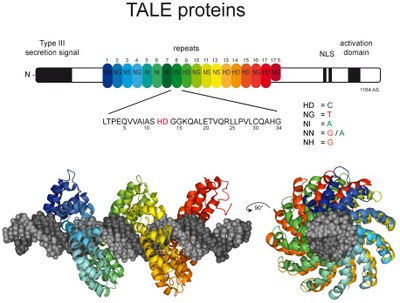
TALE nucleases consist of a TALE repeat array as DNA-binding module and the non-specific DNA-cleavage domain of the restriction endonuclease FokI as DNA-cleavage module. Because the FokI-cleavage domain itself has no further sequence specificity, off-target cleavage may occur [4]. Our group introduced an alternative design, namely to generate chimeras of a DNA recognition element (Zinc fingers or TALE arrays) and a specific restriction endonucleases such as PvuII, which showed a higher specificity in vitro [5-7]. These designer nucleases introduce a highly specific double-strand break in the genome that is either processed by homology-directed repair in the presence of a homologous repair template or by non-homologous end-joining (NHEJ) that usually results in insertions or deletions. The error-prone NHEJ can be efficiently suppressed by ‘nickases’ that produce a single-strand break rather than a double-strand break. We therefore developed a programmable site- and strand-specific nickase, by fusing a TALE array to the DNA mismatch repair protein MutH [8].
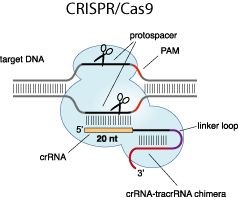
The great application potential of artificial genome targeting tools like zinc fingers, TAL effectors and CRISPR-Cas9 systems has been widely acknowledged, however, critical mechanistic details of the technologies remain largely unknown. Therefore, we now investigate the mechanism how these genome targeting tools find their specific target, as well as the thermodynamics and kinetics of DNA binding. Based on these results, we want to improve the specificity of our new designer nucleases and promote their clinical and biotechnological application.
1. Pingoud, A., Wilson, G. G. & Wende, W. (2014) Type II restriction endonucleases—a historical perspective and more, Nucleic Acids Res. 42, 7489-7527.
2. Pingoud, A. & Wende, W. (2011) Generation of novel nucleases with extended specificity by rational and combinatorial strategies, Chembiochem. 12, 1495-500.
3. Kim, H. & Kim, J.-S. (2014) A guide to genome engineering with programmable nucleases, Nat Rev Genet. 15, 321-334.
4. Gabriel, R., Lombardo, A., Arens, A., Miller, J. C., Genovese, P., Kaeppel, C., Nowrouzi, A., Bartholomae, C. C., Wang, J., Friedman, G., Holmes, M. C., Gregory, P. D., Glimm, H., Schmidt, M., Naldini, L. & von Kalle, C. (2011) An unbiased genome-wide analysis of zinc-finger nuclease specificity, Nat Biotechnol. 29, 816-23.
5. Yanik, M., Alzubi, J., Lahaye, T., Cathomen, T., Pingoud, A. & Wende, W. (2013) TALE-PvuII Fusion Proteins - Novel Tools for Gene Targeting, PLoS One. 8, e82539.
6. Schierling, B., Dannemann, N., Gabsalilow, L., Wende, W., Cathomen, T. & Pingoud, A. (2012) A novel zinc-finger nuclease platform with a sequence-specific cleavage module, Nucleic Acids Res. 40, 2623-38.
7. Fonfara, I., Curth, U., Pingoud, A. & Wende, W. (2012) Creating highly specific nucleases by fusion of active restriction endonucleases and catalytically inactive homing endonucleases, Nucleic Acids Res. 40, 847-60.
8. Gabsalilow, L., Schierling, B., Friedhoff, P., Pingoud, A. & Wende, W. (2013) Site- and strand-specific nicking of DNA by fusion proteins derived from MutH and I-SceI or TALE repeats, Nucleic Acids Res. 41, e83.
- Selected Publications of the last 5 years
-
Selected Publications of the last 5 years
1. Yanik M, Ponnam SPG, Wimmer T, Trimborn L, Müller C, Gambert I, Ginsberg J, Janise A, Domicke J, Wende W, Lorenz B, Stieger K. (2018). Development of a reporter system to explore MMEJ in the context of replacing large genomic fragments. Mol. Ther. Nucleic Acids. 11:407-415.
2. Yanik M, Müller B, Song F, Gall J, Wagner F, Wende W, Lorenz B, Stieger K. (2017). In vivo genome editing as a potential treatment strategy for inherited retinal dystrophies. Prog. Retin. Eye Res. 56:1-18.
3. Yanik, M., Alzubi, J., Lahaye, T., Cathomen, T., Pingoud, A. and Wende, W. (2013) TALE-PvuII fusion proteins – novel tools for gene targeting. PLoS One 8, e82539.
4. Heller, I., Sitters, G., Broekmans, O. D., Farge, G., Menges, C., Wende, W., Hell, S. W., Peterman, E. J. & Wuite, G. J. (2013). STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nature Methods 10, 910-916.
5. Gabsalilow, L., Schierling, B., Friedhoff, P., Pingoud, A. and Wende, W. (2013) Site- and strand-specific nicking of DNA by fusion proteins derived from MutH and I-SceI or TALE repeats. Nucleic Acids Res., 41, e83.
- Collaboration partners (alphabetic order)
-
Collaboration partners (alphabetic order)
Toni Cathomen, University Medical Center Freiburg, Germany
Thomas Lahaye, ZMBP – University of Tuebingen, Germany
Michael Kokkinidis, University of Crete, Heraklion, Greece
Elena Kubareva / Tatjana Oretskaya, Moscow State University, Russia
Ralf Seidel, University Leipzig, Germany
Knut Stieger, retinale Gentherapie - UKGM, Gießen, Germany
Gijs Wuite, VU University Amsterdam, Netherlands
