Forschungsbereiche der AG Entwicklungsbiologie der Pflanzen
ICIPS - FOR 5098
„Innovation und Koevolution in der sexuellen Reproduktion von Pflanzen“
Das Projekt...
Als die frühen Landpflanzen ihr Verbreitungsgebiet ausdehnten, setzten sie sich dem ständigen Risiko der Austrocknung aus. Daher veränderte sich auch die sexuelle Reproduktion der Pflanzen, so dass sie von Wasser unabhängiger wurden. Landpflanzen entwickelten eine ganze Reihe an wichtigen Innovationen in der sexuellen Reproduktion, wie beispielsweise die Bildung von Sporen (Pollen) die unbewegliche Spermazellen transportieren, Samenanlagen, die den reduzierten weiblichen Gametophyten (Embryosack) enthalten, und multizelluläre Embryonen, die, eingebettet in Samen, in dehydriertem Zustand effizient verbreitet werden können. Somit finden sich bei der sexuellen Fortpflanzung der Landpflanzen viele bemerkenswerte Bespiele für ganz essentielle biologischen Konzepte der „Innovation“ (z. B. der Ursprung der Samenanlage und Blüten) und der „Koevolution“ (z.B. zwischen für die Befruchtung wichtigen Signalpeptiden und deren Rezeptoren). Bisher sind die evolutionären Dynamiken und zugrundeliegenden molekularbiologischen Mechanismen dieser reproduktiven Prozesse nur rudimentär verstanden. Sieben unabhängige Projektleiter mit komplementärer Expertise aus den Bereichen Molekular-, Entwicklungs-, Zell-, und Evolutionsbiologie, unterstützt von einer Bioinformatik-Arbeitsgruppe schließen sich daher in der FOR5098 ICIPS zusammen, um erhebliche Lücken im Verständnis der molekularen Mechanismen, die Innovationen in der Evolution pflanzlicher Reproduktion ermöglichen, zu schließen. Die Projekte befassen sich (i) mit der molekularen Evolution von essentiellen Transkriptionsfaktorfamilien, den genregulatorischen Netzwerken, in die sie eingebettet sind, und (ii) mit der Evolution der Kommunikation zwischen männlichen und weiblichen Reproduktionsstrukturen. Wir werden (iii) die Grundlagen zum Vergleich zeitlicher und räumlicher Dynamik reaktiver Sauerstoffspezies (ROS) innerhalb der Evolution sexueller Reproduktion bei Landpflanzen schaffen. Der Schwerpunkt liegt auf der Analyse der molekularen Evolution und Koevolution von Genen aus Nicht-Samenpflanzen, deren Homologe essentielle morphogenetische Prozesse in Samenpflanzen, wie beispielsweise die Blüten- und Samenentwicklung sowie Befruchtungsprozesse steuern. In den Projekten der einzelnen Gruppen werden aktuellste Methoden der Molekularbiologie, Zellbiologie, in vivo Bildgebungsverfahren mit genetisch kodierten Biosensoren, Genomik, Transkriptomik, Genomeditierung und Bioinformatik in genetischen Modellorganismen innerhalb der Nicht-Samenpflanzen und Blütenpflanzen kombiniert. Die entsprechenden Methoden stehen für diese Pflanzenarten, darunter das Lebermoos Marchantia, das Laubmoss Physcomitrium und der Farn Ceratopteris erst seit kurzer Zeit zur Verfügung. Weiterhin will die FOR ICIPS Wissenschaftler/innen und Expertise im Bereich der pflanzlichen evolutionären Entwicklungs- und Reproduktionsbiologie zusammenführen um bereits etablierte Methoden an neuen, Nicht-Samenpflanzen Modellarten zur Anwendung zu bringen.
Koordinator: Dr. Romain Scalone
Funding: 2021-2025 German Research Foundation (DFG)
Evolution von Gennetzwerken: Die Ranunculales als Modellordnung für evolutionäre Innovation
Wie Veränderungen in regulatorischen Gennetzwerken (GRNs) den Ursprung neuer, complexer Merkmalen bedingen, oder, kurzgefasst, „Wie entsteht Neues in der Evolution?“ ist eine zentrale Frage der modernen Evolutionsbiologie. Wir möchten diesen molekulargenetischen Ursprung neuer Merkmale am Modell der Ordnung Ranunculales (Hahnenfußgewächse) untersuchen. Ranunculales sind recht ursprüngliche Eudikotyle (Zweikeimblättrige Pflanzen) und umfassen morphologisch sehr diverse Arten wie Hahnenfuß, Mohn, Akelei oder Rittersporn. Ranuncuales sind deshalb besonders, weil mehrere morphologische Merkmaler mehrfach unabhängig voneinander entstanden sind, z.B. gespornte Blütenorgane, Zygomorphie oder die Reduktion von Perianthorganen. Darüber hinaus sind in dieser Blütenpflanzenordnung neue Blütenorgane, Reduktion von gewirtelter zur spiraligen Phyllotaxy, Diözie, und Windbestäubung aufgetaucht. Viele Arten innerhalb der Ranunculales können mit Virus-induziertem Gensilencing behandelt
werden, wodurch Gene gezielt ausgeschaltet werden können, um deren Funktion in späteren Stadien dieses Projektes untersuchen zu können. Wir schlagen vor, 20 Transkriptome von sieben Ranunculales Arten zu sequenzieren (Eschscholzia californica, Capnoides sempervirens, Pteridophyllum canadensis, Thalictrum thalictroides, Aquilegia coerulea, Nigella damascena, Staphisagria picta), die diese evolutionären Innovationen zeigen. Die Transkriptome werden von z.B. Knospen unterschiedlicher Stadien, verschiedener Blütenorgane, Zeitserien von Petalengewebe und vegetativem Gewebe stammen. Zudem planen wir Genomsequenzierungen der vier Arten, deren Genome noch nicht sequenziert wurden, um die Transkriptomanalysen mittels Transkript-Identifizierung und -Mapping zu optimieren. Auf lange Sicht beabsichtigen wir die Identifikation von Co- Expressionsgennetzwerken und wollen den Grad der Konservierung von co-exprimierten Teilen von GRNs beurteilen und die Teile identifizieren, die sich zwischen den Ranunculales-Arten unterschieden, um sie mit den entsprechenden evolutionären Innovationen zu korrelieren. Das Ziel dieses Projektes ist es, die nötigen Resourcen zur Verfügung zu stellen, um vergleichende Transkriptom- und Genomanalysen in Ranunculales durchführen zu können. Diese sind die Voraussetzung, um die molekularen Grundlagen des Ursprungs neuer morphologischer Merkmale zu verstehen.
The evolution of plasmodesmata 
Plant cells are interconnected by plasmodesmata (PD), i.e. complex cytoplasmic channels that mediate intercellular communication and symplasmic exchange of (macro)molecules. The invention of PD was a crucial step in plant evolution, since it allowed i) the development of increasingly complex plant bodies with division of labour between spatially patterned tissues and organs and it enabled ii) coordinated strategies to face the hazardous biotic and abiotic stresses occurring during the conquest of land.
In seed plants, PD networks are highly dynamic and undergo drastic functional and structural changes controlling developmental processes, metabolic acclimatisation and pathogen responses. Yet, information on PD networks in non-seed plants and streptophyte algae is scarce and often inconsistent. Within the streptophytes, homology of PD-like structures in the ZCC grade algae and land plant PD is hypothesised, although there are still uncertainties pertaining to a uniform PD architecture and a common mode of PD formation which will be addressed in the present project.
Postcytokinetic formation of secondary PD in pre-existing cell walls is an appropriate mode to adjust PD numbers (and transport capacities) to changing requirements and has evolved (independently?) at least in some lycophyte lineages and spermatophytes. Whether secondary PD formation also occurs in other land plant lineages is unclear and will be investigated in the project.
Since the project is part of the DFG priority programme 2237 “MAdLand - Molecular Adaptation to Land: plant evolution to change”, we will perform our microscopic analyses on the MAdLand model plant species which represent distinct taxa of streptophyte algae and non-seed land plants. We aim to discover basic differences in PD structure and origin among the streptophyte lineages, the molecular basis of which will be addressed in the second funding period.
Funding: 2020-2023 German research Foundation (DFG)
Cooperation partners: Stefan Rensing, University of Marburg (MAdLand coordinator), cf. https://madland.science/ and http://madland.science/projects.php
Phylotranscriptomics of the carpel developmental toolkit - an evodevo study towards understanding the origin of flowering plants
We know relatively little about the origin of angiosperms, a group of plants that dominate most terrestrial ecosystems and pr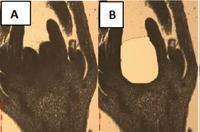
The general aim of this project is to identify a minimal set of genes required for carpel development in angiosperms to understand which genetic prerequisites were required to build the ancestral carpel, and thus, how a crucial step during angiosperm origin was completed. Subsequently, the molecular functions and genetic interactions of this set of genes will be identified to predict the ancestral state of the carpel development network and to learn which genes carried out which functions in the ancestral angiosperm. This project is the first to systematically analyze carpel developmental regulators in an unbiased way across a wide range of phylogenetically important taxa by a combination of laser capture microdissection and phylotranscriptomics. With the newly published genomes of A. trichopoda and P. abies and the availability of the W. mirabilis transcriptomes as second gymnosperm from a divergent lineage, this project is now a timely and realistic undertaking.
Funding: 2016 – 2019 German Research Foundation (DFG)
Cooperation partners: Alexander Goesmann and Oliver Rupp, Institute for Bioinformatics and Systems Biology, JLU Gießen, Knut Beuerlein, Rudolph-Buchheim-Institute for Pharmacology, JLU Gießen
Contact: Annette Becker
Evolutionary genetics of carpel development using California poppy (Eschscholzia californica) as a new model species
All flowering plants have carpels, female reproductive structures that enclose the eggs and subsequently develop into seedpods and fruits. Carpel development genes are being defined in the model species Arabidopsis, an advanced flowering plant. But the evolutionary origin of carpels is not clear. In this project, we will identify and characterize genes that control carpel development in a more primitive plant, California poppy (Eschscholzia californica). This new model species is a basal eudicot that can be manipulated transgenically. Comparison of carpel genes in poppy and Arabidopsis will help reveal core genes that underlie carpel development in all flowering plants. This will allow us to identify genes that play the same important role in carpel development across dicots, and also those that play more specialized roles. The underlying question is how gene networks, consisting of developmental genes from different gene families, govern plant development and how these networks evolve resulting in changes to plant structure.

Funding: 2005-2014 German Research Foundation (DFG), continuation with JLU funding
Contact: Annette Becker
Gefährdungsbeurteilung der AG Entwicklungsbiologie der Pflanzen
