Prof. Dr. Rainer Renkawitz
Prof Dr. R. Renkawitz
Institut für Genetik Justus-Liebig-Universität Gießen Heinrich-Buff-Ring 58-62 35392 Gießen
|
Forschung:
Epigenetische Mechanismen zur Kontrolle der Genaktivität
Der größte Teil der 25 000 Gene im Genom der Säuger ist in vielen Zelltypen und Geweben inaktiv. Innerhalb einer Population von Zellen bleiben die meisten dieser Gene inaktiv und vererben ihren epigenetischen Zustand auf die Tochterzellen. Der Erhalt des abgeschalteten Zustands ist eine Grundvoraussetzung für die Embryonalentwicklung, für Zelldifferenzierung und für die normale Funktion der Gewebe. Genom-weite, epigenetische DNA- und Chromatin-Modifikation spielen eine grundlegende Rolle in der Regulation der Genaktivität.
Häufig ist die Veränderung des epigenetischen Musters in Krankheiten des Menschen, einschließlich der Tumorentstehung, involviert. Tumor-Suppressor-Gene sind fälschlich abgeschaltet, während Proto-Onkogene aktiviert sind. Daher findet man bei der Tumorentstehung beide Situationen, sowohl Hypermethylierung als auch Hypomethylierung genomischer DNA. Zurzeit kennt man bereits für sehr viele menschliche Erkrankungen eine ursächliche genetische Veränderung. Trotz dieses Wissens bleibt die grundsätzliche Frage auch heute noch unbeantwortet, wie die molekulare Repression der Genaktivität vermittelt wird und wie sie von einer Zellteilung zur nächsten erhalten bleibt. Die generellen Mechanismen schließen das sogenannte "Chromatin Remodelling", Histon-Modifikationen und DNA-Methylierung ein. Die molekulare Funktion dieser Mechanismen im Zusammenhang von Gen-Repression, Gen-Aktivierung, Gen-Isolation, sowie der subnukleären Verteilung der Gene werden in unserer Arbeitsgruppe detailliert untersucht.
Forschungsschwerpunkte
|
|
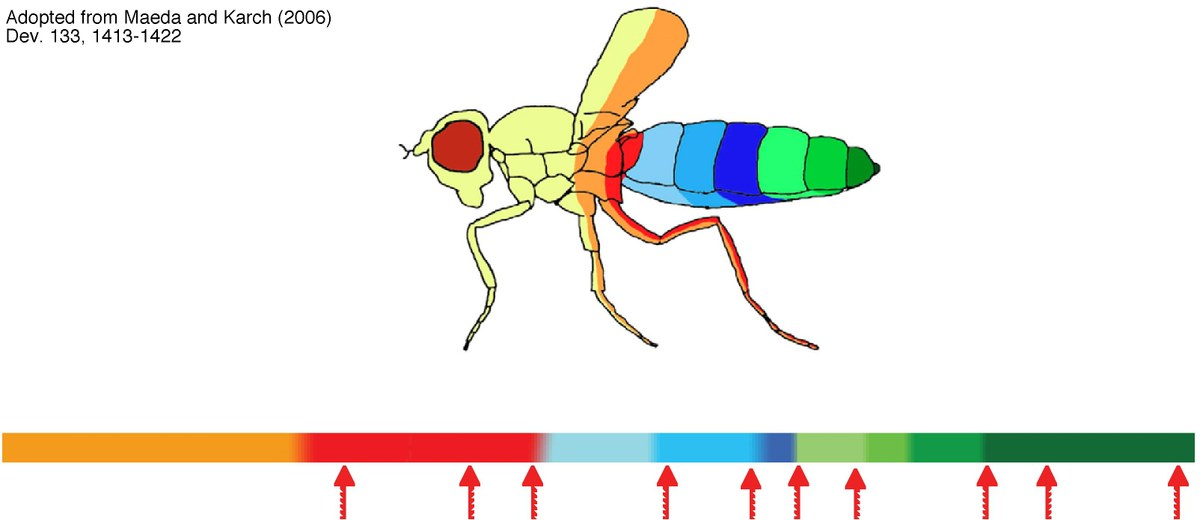
Key mediators of the enhancer blocker function of Drosophila CTCFFunctional eukaryotic gene units including all of their regulatory elements are insulated from each other to allow the action of regulatory elements on the appropriate gene. We and others have analyzed the DNA binding factor CTCF, which is the only protein known to mediate a directional blocking activity of enhancer elements in vertebrates (3, 8). In focusing on Drosophila CTCF, we found out that dCTCF mediates enhancer blocking on the Fab-8 insulator and binds to several other known insulator sequences (1, 2, 9). One set of dCTCF binding sites separates the homeotic genes responsible for the segmental identity in flies (2, 5). Deletion of dCTCF causes a defect in abdominal segmentation (homeotic mutation) and lethality (1). Search for co-factors involved in dCTCF function revealed a new, unknown protein (7), several proteins known to mediate chromatin remodelling (4, 5, 6) and chromatin modification (10). These results support a model for insulator function: DNA bound dCTCF recruites several co-factors removing changes in chromatin modification, which may have spread from flanking regions.
|
|
1. Mohan, M., Bartkuhn, M., Herold, M., Philippen, A., Heinl, N., Bardenhagen, I., Leers, L., White, R.A.H., Renkawitz-Pohl, R., Saumweber, H. and Renkawitz, R. (2007). The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26, 4203-4214. 2. Bartkuhn, M., Straub, T., Herold, M., Herrmann, M., Rathke, C., Saumweber, H., Gilfillan, G.D., Becker, P.B. and Renkawitz, R. (2009). Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 28, 877-888. 3. Herold, M., Bartkuhn, M. and Renkawitz, R. (2012) CTCF: Insights into insulator function during development. Development 139, 1045-1057. 4. Ahanger, S.H., Günther, K., Weth, O., Bartkuhn, M., Bhonde, R.R., Shouche, Y.S., and Renkawitz, R. (2014). Ectopically tethered CP190 induces large-scale chromatin decondensation. Scientific Reports 4, 3917. 5. Bohla, D.*, Herold, M*, Panzer, I, Buxa, M.K., Ali, T., Demmers, J., Krüger, M., Scharfe, M., Jarek, M., Bartkuhn, M.1, Renkawitz, R. (2014). A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking. PLoS One 9: e107765. 6. Weth, O., Paprotka, C., Günther, K., Schulte, A., Baierl, M., Leers, J., Galjart, N. and Renkawitz, R. (2014). CTCF Induces Histone Variant Incorporation, Erases the H3K27me3 Histone Mark and Opens Chromatin. Nucl. Acids Res. 42, 11941-11951. 7. Maksimenko, O.*, Bartkuhn, M.*, Stakhov, V.*, Herold, M.*, Zolotarev, N., Jox, T., Buxa, M.K., Kirsch, R., Bonchuk, A., Fedotova, A., Kyrchanova, O., Renkawitz, R., Georgiev, P. (2015) Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25, 89-99. 8. Ali, T., Renkawitz, R. and Bartkuhn, M. (2016). Insulators and domains of gene expression. Curr Opin Genet Dev 37, 17-26. 9. Buxa, M.K., Slotman, J.A., van Royen, M.E., Paul, M.W., Houtsmuller, A.B. and Renkawitz, R. (2016). Insulator speckles associated with long-distance chromatin contacts. Biology Open 5, 1266-1274. 10. Ali, T., Krüger, M., Bhuju, S., Jarek, M., Bartkuhn, M. and Renkawitz, R. (2017). Chromatin binding of Gcn5 in Drosophila is largely mediated by CP190. Nucl. Acids Res. 45, 2384-2395.
|
|
|
|
Role of multi zinc finger proteins in the dynamic change of the nuclear architecture during cell cycle and differentiation
The multi-zinc finger proteins CTCF and CTCFL are unique and conserved factors with a role in transcriptional regulation, the organization of chromatin into distinct domains and imprinting (2, 4). CTCF is ubiquitously expressed with cell-specific and gene-specific functions (3, 5, 6, 7). In contrast, the related factor CTCFL is expressed in the testis (8). Abnormal up-regulation of CTCFL, on the other hand, may be linked to tumorigenesis. Thus, while binding to similar sites in the genome, these proteins do have distinct roles. We could show that CTCF, in contrast to CTCFL and to other zinc finger proteins, remains bound to mitotic chromatin (1). Despite very similar zinc finger domains of both proteins, they differ in their molecular function. Specificity of chromatin binding is driven by nucleosomal modification and density (8). CTCF is actively opening compact chromatin, whereas CTCFL cannot (9).
|
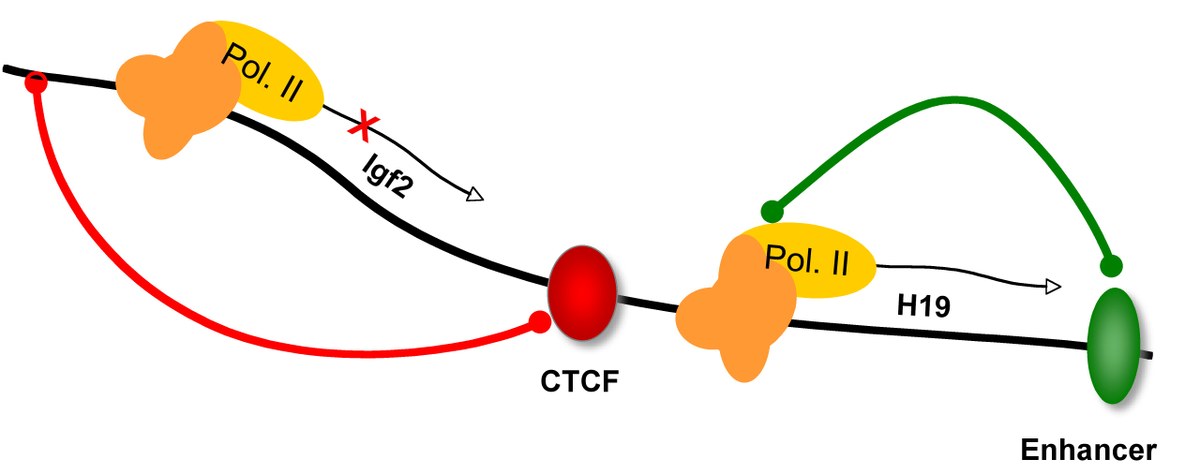
| 1. Burke, LJ., Zhang, R., Bartkuhn, M., Tiwari, VK., Tavoosidana, G., Kurukuti, S., Weth, C., Leers, J., Galjart, N., Ohlsson, R., and Renkawitz, R. (2005) CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 24, 3291-3300.
2. Bartkuhn, M. and Renkawitz, R. (2008) Long range chromatin interactions involved in gene regulation. Biochim. Biophys. Acta 1783, 2161-2166. 3. Heath, H., de Almeida, C.R., Sleutels, F., Dingjan, G., van de Nobelen, S., Jonkers, I., Ling, K.W., Gribnau, J., Renkawitz, R., Grosveld, F., Hendriks, R.W. and Galjart, N. (2008). CTCF regulates cell cycle progression of αβ T cells in the thymus. EMBO J. 27, 2839-2850. 4. Ohlsson, R., Bartkuhn, M. and Renkawitz, R. (2010). CTCF shapes chromatin by multiple mechanisms. Chromosoma 119, 351-360. 5. Weth, O., Weth, C., Bartkuhn, M., Leers, J., Uhle, F. and Renkawitz, R. (2010) Modular Insulators: Genome wide search for composite CTCF / Thyroid hormone receptor binding sites. PLoS ONE 5, e10119. 6. van de Nobelen, S., Rosa-Garrido, M., Leers, J., Heath1, H., Soochit, W., Joosen, L., Jonkers, I., Demmers, J., van der Reijden, M., Torrano, V., Grosveld, F., Delgado, M.D., Renkawitz, R., Galjart, N. and Sleutels F. (2010) CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics & Chromatin 3, 19. 7. Weth, O. and Renkawitz, R. (2011) CTCF function is modulated by neighboring DNA binding factors. Biochem. Cell Biol. 89, 459-468. 8. Sleutels, F., W. Soochit, M. Bartkuhn, H. Heath, S. Dienstbach, P. Bergmaier, V. Franke, M. Rosa-Garrido, S. van de Nobelen, L. Caesar, M. van der Reijden, J. C. Bryne, W. van Ijcken, J. A. Grootegoed, M. D. Delgado, B. Lenhard, R. Renkawitz, F. Grosveld and N. Galjart (2012). The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics & Chromatin 5(1): 8. 9. Weth, O., Paprotka, C., Günther, K., Schulte, A., Baierl, M., Leers, J., Galjart, N. and Renkawitz, R. (2014). CTCF Induces Histone Variant Incorporation, Erases the H3K27me3 Histone Mark and Opens Chromatin. Nucl. Acids Res. 42, 11941-11951 |
|