The world of small RNAs in Alphaproteobacteria
Sudden changes in the environment entail the need for sophisticated regulation of cellular responses in order to adapt and survive. Bacteria regulate their gene expression on both transcriptional and post-transcriptional level. Besides intrinsic regulatory elements of mRNAs, like riboswitches, small regulatory RNAs (sRNAs) are at the forefront of post-transcriptional regulation. Typical trans-encoded sRNAs influence their target mRNAs by means of stability and/or translation initiation. They are commonly induced upon stress and involved in regulatory fine-tuning and feedback regulations.
In the group of Alphaproteobacteria metabolic diversity is enormous and even in a single species high metabolic flexibility can exist. Environmental cues directly determine the bacterial life-style/metabolism and the question of how these bacteria regulate their gene expression is of certain interest.
Small RNAs in Rhodobacter sphaeroides
The purple bacterium Rhodobacter sphaeroides is a well-studied model organism with regard to the regulation of photosynthesis gene expression and the response to oxidative and photooxidative stress. The search for sRNAs using a differential RNA-sequencing approach (dRNA-seq) revealed that many of these small regulators exist in R. sphaeroides and implied their regulatory relevance (Figure 1). The functional characterization of these sRNAs is currently the main focus. We found one sRNA (RSs2430) that is controlled by the oxygen-dependent two component system PrrB/PrrA and is involved in regulation under oxygen depletion. The RSs0019 sRNA specifically responds to photooxidative stress caused by singlet oxygen in an RpoE-dependent manner and may fine-tune sulfur metabolism. Interestingly, RSs0019 contains a small open reading frame (ORF) and might therefore represent a dual-function RNA with a regulatory and a coding function. Three additional sRNAs (RSs0680a, RSs1543 and RSs2461) belong to the RpoHI/RpoHII regulon and most likely depict RNA regulators with a role under more general stress conditions. Last but not least, the strongly abundant sRNA RSs0682 undergoes a singlet oxygen-specific processing which depends on the important RNA-binding protein Hfq. We successfully over-expressed several of these sRNAs, resulting in distinct phenotypes and altered global expression patterns as analyzed by microarray and proteome studies. The bioinformatic prediction of interaction sites of sRNA/mRNA pairs confines the set of direct mRNA targets, which are subsequently tested by in vivo and in vitro studies. In addition, the important RNA-binding protein Hfq was demonstrated to have a global regulatory function in R. sphaeroides and to interact with several of the above mentioned sRNAs (Figure 1). We therefore study the role of Hfq in sRNA function in R. sphaeroides.
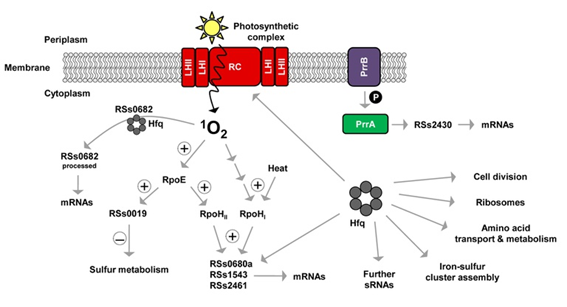
Figure 1: Model of sRNAs and Hfq in Rhodobacter sphaeroides.
Role of an sRNA repeat in the general stress response of Alphaproteobacteria
In R. sphaeroides a repeat of four homologous sRNAs was identified by dRNA-seq. The sRNAs RSs0680a-d are induced from an RpoHI/RpoHII dependent promoter and co-transcribed with RSP_6037, encoding a hypothetical protein with a domain of unknown function (DUF) 1127 (Figure 2A). The sRNAs fold into a typical secondary structure containing two stem-loops with the diagnostic CCUCCUCCC motif within the loops (Figure 2B). We are interested in the regulatory mechanisms and the role in the general stress response of these sRNAs. They are conserved among Alphaproteobacteria and regularly associated with DUF 1127 proteins. We therefore investigate this set of sRNAs in R. sphaeroides and related Alphaproteobacteria, as e.g. rhizobia.
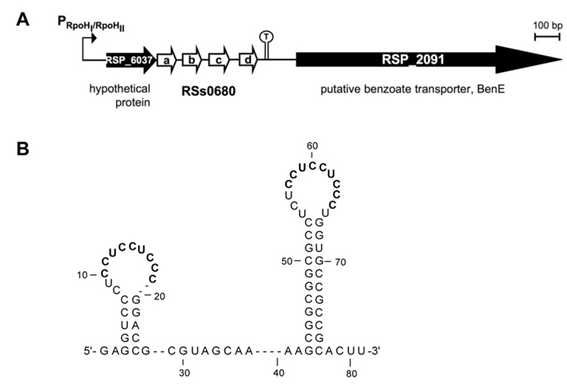
Figure 2: The homologous RSs0680 sRNAs. (A) Genetic localization of RSP_6037 and RSs0680a-d. (B) Consensus secondary structure of RSs0680-type sRNAs containing the CCUCCUCCC motives.
Small RNAs in rhizobia
Using a comparative genomics approach, candidates for small RNAs were predicted and verified in the plant symbionts Sinorhizobium meliloti and Bradyrhizobium japonicum. We analyzed several growth phase dependent small RNAs of S. meliloti, the nitrogen fixing symbiont of alfalfa. These sRNAs are conserved and expressed in representatives of the genera Sinorhizobium and Rhizobium. Two of them are Hfq-dependent: Hfq is important for their stability and degradation pathway in S. meliloti. The expression of sRNAs in B. japonicum, the symbiont of soyben plants, was studied under free-living conditions and in root nodules. Northern blots, microarrays and deep sequencing analyses revealed differential expression of several sRNAs under the two physiological conditions. Based on the deep sequencing data, we are currently mapping sRNAs, their transcriptional start sites and putative promoters. Future analyses aim to unravel the role of sRNAs in the interaction between bacteria and plants. The projects are performed in cooperation with the research groups of A. Becker (Marburg), M. Gelfand (Moskau), H.-M. Fischer (ETH Zürich), W.R. Hess (Freiburg), J. Vogel and C. Sharma (Würzburg).
Methods
Cultivation of Alphaproteobacteria; Cloning procedures; RNA isolation; Northern blotting; real time RT-PCR; Microarray; RNA 5’ and 3’ end detection (primer extension, RNA protection, 5’ and 3’ RACE); EMSA; In vitro transcription and in vitro RNA degradation; Isolation and characterization of recombinant proteins; Western blotting; 2D gel electrophoresis; in vivo reporter studies
Reference
Grützner J, Remes B, Eisenhardt KMH, Scheller D, Kretz J, Madhugiri R, McIntosh M, Klug G. sRNA-mediated RNA processing regulates bacterial cell division. Nucleic Acids Res. 2021 Jul 9;49(12):7035-7052. doi: 10.1093/nar/gkab491. PMID: 34125915; PMCID: PMC8266604
Reuscher CM, Klug G. Antisense RNA asPcrL regulates expression of photosynthesis genes in Rhodobacter sphaeroides by promoting RNase III-dependent turn-over of puf mRNA. RNA Biol. 2021 Jan 11:1-13. doi: 10.1080/15476286.2020.1857520.
Spanka DT, Klug G. Maturation of UTR-Derived sRNAs Is Modulated during Adaptation to Different Growth Conditions. Int J Mol Sci. 2021 Nov 12;22(22):12260. doi: 10.3390/ijms222212260. PMID: 34830143; PMCID: PMC8625941.
Förstner KU, Reuscher CM, Haberzettl K, Weber L, Klug G. RNase E cleavage shapes the transcriptome of Rhodobacter sphaeroides and strongly impacts phototrophic growth. Life Science Alliance 2018 Aug. Vol. 1, No. 4. doi: 10.26508/lsa.201800080
Eisenhardt K, Reuscher C, Klug G. PcrX, an sRNA derived from the 3'- UTR of the Rhodobacter sphaeroides puf operon modulates expression of puf genes encoding proteins of the bacterial photosynthetic apparatus., Mol Microbiol. 2018 Jul 11. doi: 10.1111/mmi.14076.
Weber L, Thoelken C, Volk M, Remes B, Lechner M, Klug G. The Conserved Dcw Gene Cluster of R. sphaeroides Is Preceded by an Uncommonly Extended 5' Leader Featuring the sRNA UpsM. PLoS One. 2016 Nov 1;11(11)
Müller KM, Berghoff BA, Eisenhardt BD, Remes B, Klug G. Characteristics of Pos19 - A Small Coding RNA in the Oxidative Stress Response of Rhodobacter sphaeroides. PLoS One. 2016 Sep 26;11(9):e0163425. doi: 10.1371/journal.pone.0163425. eCollection 2016.
Peng T, Berghoff BA, Oh JI, Weber L, Schirmer J, Schwarz J, Glaeser J, Klug G. Regulation of a polyamine transporter by the conserved 3' UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides.RNA Biol. 2016 Oct 2;13(10):988-999
Adnan, F., Weber, L., Klug, G. (2015) The sRNA SorY confers resistance during photooxidative stress by affecting a metabolite transporter in Rhodobacter sphaeroides. RNA Biol. Apr 1:0. [Epub ahead of print]
Billenkamp, F., Peng, T., Berghoff, B.A., Klug, G. (2015) A cluster of four homologous small RNAs modulates C1 metabolism and the pyruvate dehydrogenase complex in Rhodobacter sphaeroides under various stress conditions. J. Bacteriol. 197: 1839-1852
Damm, K., Bach, S., Müller, K.M., Klug, G. Burenina, O.Y., Kubareva, E.A. Grünweller, A., Hartmann, R. (2015) Improved northern blot detection of small RNAs using EDC crosslinking and DNA/LNA probes. Methods Mol. Biol. 1296:41-51
Damm, K., Bach, S., Müller, K.M., Klug, G., Burenina, O.Y., Kubareva, E.A. Grünweller, A., Hartmann, R. (2015) Impact of RNA isolation protocols on RNA detection by northern blotting. Methods Mol. Biol. 1296:29-38
Hess, W., Berghoff, B., Wilde, A., Steglich, C., Klug, G. (2014) Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biology 11: 413-426
Mank, N. N., Berghoff, B. A., Klug, G. (2013) A mixed incoherent feed-forward loop contributes to the regulation of bacterial photosynthesis genes. RNA Biol. 2013 Mar;10(3):347-52
Mank,N. N., Berghoff, B. A., Hermanns, Y. N., Klug, G. (2012) Regulation of bacterial photosynthesis genes by small non-coding RNA PcrZ. Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):16306-11. doi: 10.1073/pnas.1207067109. Epub 2012 Sep 17.
Madhugiri R, Pessi G, Voss B, Hahn J, Sharma CM, Reinhardt R, Vogel J, Hess WR, Fischer HM, Evguenieva-Hackenberg E. (2012) Small RNAs of the Bradyrhizobium/Rhodopseudomonas lineage and their analysis. RNA Biol. 2012 Jan 1;9(1). [Epub ahead of print]
Glaeser, J., Nuss, A.M., Berghoff, B.A., Klug, G. (2011) Singlet oxygen stress in microorganisms. In R. Poole, R.K. (ed): Adv. in Microb. Physiol. Vol. 58 (Amsterdam), pp. 141-173
Berghoff, B.A., Glaeser, J., Sharma, C.M., Zobawa, M., Lottspeich, F., Vogel, J., Klug, G. (2011) Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol. Microbiol. 80: 1479-1495
Berghoff, B.A., Glaeser, J., Nuss, A.N., Zobawa, M., Lottspeich, F., Klug, G. (2011) Anoxygenic photosynthesis and photooxidative stress: A particular challenge for Roseobacter. Environm. Microbiol.
Nuss, A.M., Glaeser, J., Berghoff, B.A., Klug, G. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J Bacteriol 2010 192:2613-23
Schlüter JP1, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Jänicke S, Becker JD, Giegerich R, Becker A. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010 Apr 17;11:245. doi: 10.1186/1471-2164-11-245.
Madhugiri R, Evguenieva-Hackenberg E. RNase J is involved in the 5'-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett. 2009 Jul 21;583(14):2339-42. doi: 10.1016/j.febslet.2009.06.026. Epub 2009 Jun 21.
Voss B, Hölscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, Becker A, Evguenieva-Hackenberg E. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti. Biochem Biophys Res Commun. 2009 Dec 11;390(2):331-6. doi: 10.1016/j.bbrc.2009.09.125. Epub 2009 Oct 2.
Berghoff, B.A., Glaeser, J., Sharma, C., Jörg Vogel, J., Klug, G. Photooxidative stress induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol. 2009 74:1497-512.
Glaeser J, Zobawa M, Lottspeich F, Klug G. Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter. J Proteome Res. 2007 Jul; 6:2460-71
