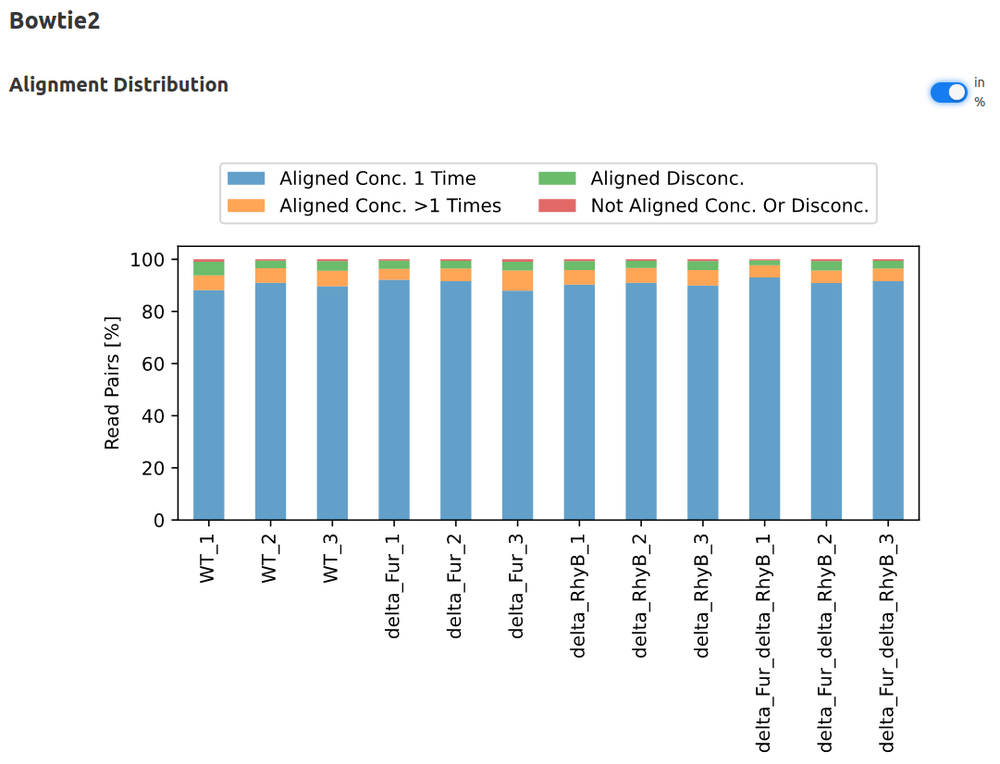
Curare
Curare is a freely available analysis pipeline for reproducible, high-throughput, bacterial RNA-Seq experiments. Define standardized pipelines customized for your specific workflow without the necessity of installing all the tools by yourself.
Curare is implemented in Python and uses the power of Snakemake and Conda to build and execute the defined workflows. Its modulized structure and the simplicity of Snakemake enables developers to create new and advanced workflow steps.
Features
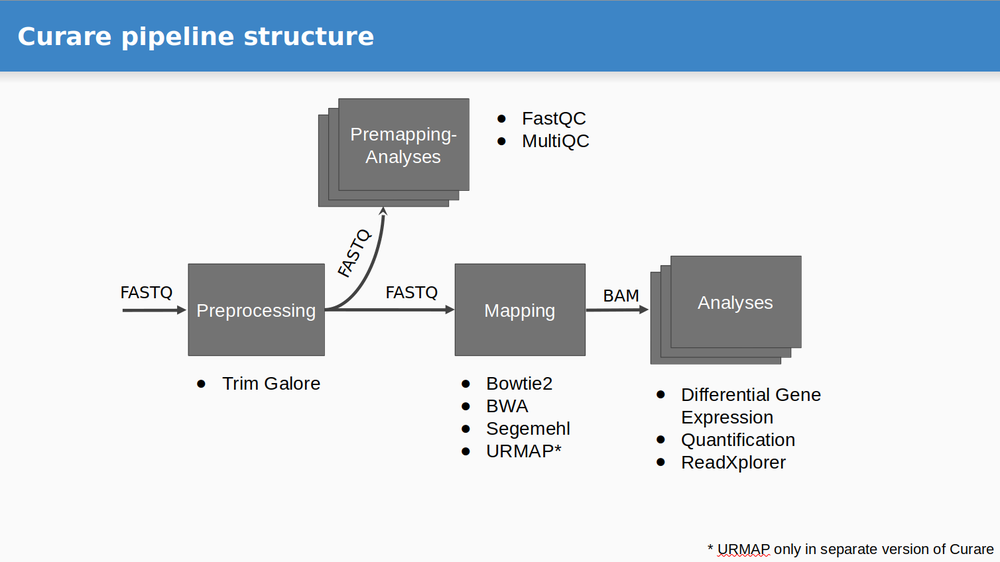
Curare was developed to simplify the automized execution of RNA-Seq workflows on large datasets. Each workflow can be divided into four steps: Preprocessing, Premapping, Mapping, and Analysis.
Currently available modules
- Preprocessing
- Trim-Galore
- Premapping
- FastQC
- MultiQC
- Mapping
- Bowtie
- Bowtie2
- BWA (Backtrack)
- BWA (Mem)
- BWA (SW)
- Segemehl
- Star
- Analysis
- Count Table (FeatureCounts)
- Differential Gene Expression Analysis (DESeq2)
- Normalized Coverage
- ReadXplorer
For further information: https://github.com/pblumenkamp/Curare