RNase as a component of inducible gene expression
|
Project 1: RNase as a component of inducible gene expression
|
   |
|
Model of the RNase NynA from Sinorhizobium meliloti
The protein components of the RNase complex are arranged together to achieve maximal degradation of RNA, preventing gene expression. When a specific chemical is supplied, RNase activity is inhibited, allowing gene expression. © Vasundra Srinivasan
|
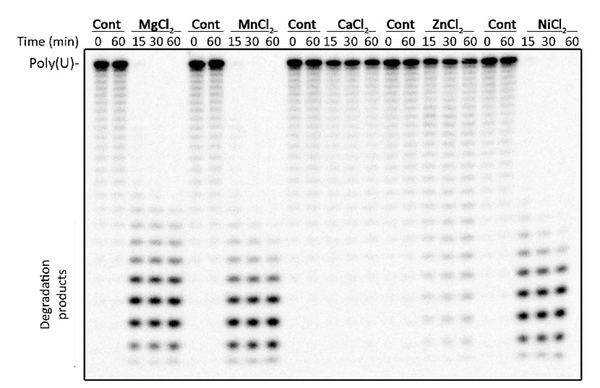
Most inducible gene expression systems are controlled at the level of transcription. Transcription is a process in gene expression in which information encoded in the DNA is converted to RNA. A later step in gene expression is translation, in which the information in the RNA is converted to protein, the most common ‘workhorse’ of the cell. Unlike transcription, translation has rarely been controllable via inducers. One of my projects is focused on the use of RNases, which are specialized proteins that rapidly cut and degrade RNA, often in the presence of one or more metal ions. In this project, a novel RNase has been found whose activity can be additionally controlled by a specific hormone. The unusual ability of this RNase to respond to chemical inducers will be harnessed as a method for controlling gene expression. If successful, this RNase will be the first of its kind to be included in a system which involves control over both transcription and translation for simultaneously fine-tuning gene expression. |