Projects
Research topics
We are investigating regulation at the level of RNA in bacteria, particularly in the early response to antibiotc exposure. We are also interested in very small proteins translated from own small ORFs. we are using rhizobia and E. coli to study posttranscriptional gene regulation by the trp attenuator, SAM-II riboswitches and regulatory upstream ORFs.
The small proteome in rhizobia
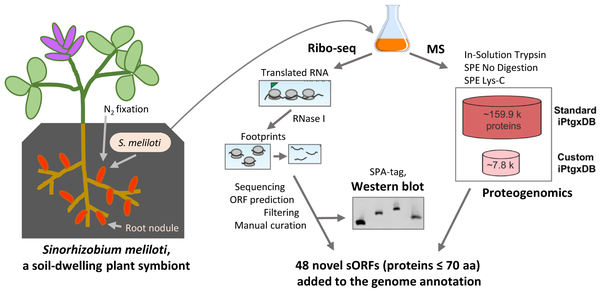
Detection of small ORFs and small proteins in Sinorhizobium meliloti.
From Hadjeras et al. (2022), https://doi.org/10.1093/femsml/uqad012
Small proteins are often neglected and are missing in genome annotations, although they can have important functions [1]. Recently, we identified novel small ORFs by ribosome probiling, mass spectrometry and western blot analysis [2]. The data of this Ribo-seq analysis, the first of a Hyphomicrobiales member, can be viewed with an interactive online JBrowse instance (http://www.bioinf.uni-freiburg.de/ribobase).
In the past, we mapped genome-wide transcription start sites and predicted promoters in the reannotated genome of the soybean symbiont Bradyrhizobium japonicum (Bradyrhizobium diazoefficiens). This analysis led to the discovery and annotation of many small ORFs [3-5].
The functions of small ORFs and small proteins are in the focus of our research.
SAM homeostasis in S. meliloti
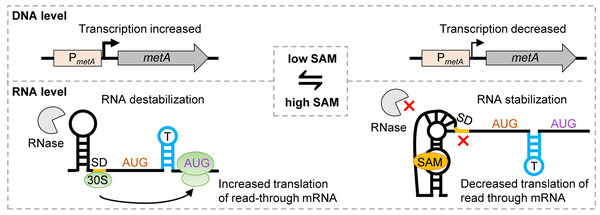
Model of metA regulation. On the left: Under low SAM conditions, transcription from the metA promoter (PmetA) is increased and the riboswitch is in the SAM-free state. The Shine-Dalgarno (SD) sequence downstream of the riboswitch aptamer is accessible and activates translation in the read-through mRNA from the starting codon AUG2. Under these conditions, metA mRNA is destabilized. On the right: Under high SAM conditions, transcription from PmetA is decreased, the riboswitch is in the SAM-bound state, the SD controlled by the riboswitch is not accessible and metA translation is repressed. Under these conditions, metA mRNA is stabilized.
From Scheuer et al (2002), doi: 10.1080/15476286.2022.2110380.
Riboswitches are cis-acting RNA elements, which specifically bind a metabolite and regulate the expression of the downstream genes [6]. SAM riboswitches interact with the S-adenosyl-L-methionine (SAM), the main methyl donor for biochemical reactions in the cell. S. meliloti harbors two SAM-II riboswitches, which are located upstream of the methionine biosynthesis genes metA and metZ and were predicted to regulate the transcription of these genes. Recently, we found two unexpected functions of the metA riboswitch: in the SAM-bound state, it downregulates metA translation by a novel mechanism, and additionally stablizes the mRNA. This probably helps to adapt to changing conditions and maintain SAM homeostasis [7]. Additional putative functions of the SAM-II riboswitch in S. meliloti are under investigation.
In the past, we have shown that in S. meliloti, the ribonucleases RNase E and RNase J are important for the SAM homeostasis [8].
The trp attenuator
In prokaryotes, transcription and translation can be coupled. Therefore, in bacteria efficiency of translation of small upstream open reading frames (uORFs) can determine whether the downstream genes are transcribed. This mechanism depends on mutually exclusive RNA secondary structures and is known as ribosome-dependent transcription attenuation. Many Gram-negative bacteria have transcription attenuators upstream of amino acid (aa) biosynthesis operons. A prime example is the trpEDCBA operon in Escherichia coli. Its mRNA leader harbors the uORF trpL, which contains two consecutive tryptophan (Trp) codons. Under conditions of Trp shortage, ribosome pausing at the Trp codons prevents the formation of a transcriptional terminator between trpL and trpE. When Trp is available, trpL is efficiently translated (and thus the so called leader peptide is produced) and a small attenuator RNA (which harbors trpL) is generated [9].
Usually attenuator RNAs and leader peptides are considered non-functional. Our work suggests that the leader peptide peTrpL and the small RNA attenuator (sRNA) rnTrpL are functional in trans. We also found that the trp attenuator responds not only to tryptophan availability, but also to translation inhibition by antibiotics.
The conserved attenuator sRNA rnTrpL: a riboregulator responding to nutrient availability
In contrast to E. coli, S. meliloti harbors three trp operons and only one of them, trpE(G), is regulated by transcription attenuation. The trpE(G) operon is not regulated by a transcriptional repressor and is constitutively transcribed during growth. When enough Trp is available, trpE(G) transcription is abolished and the sRNA rnTrpL is generated. This sRNA base-pairs with trpD in the polycistronic trpDC mRNA and destabilizes the transcript [10]. Additionally, it downregulates the quorum sensing autoinducer synthase gene sinI [11].
In E. coli at low Trp concentration, transcription of the trpEDCBA operon is derepressed and its expression is regulated by attenuation. Under conditions of Trp sufficiency, the attenuator sRNA rnTrpL is produced [12]. We have shown that rnTrpL directly downregulates expression of dnaA, which encodes the master regulator of initiation of chromosome replication. Thus, the sRNA rnTrpL links the availability of Trp, which is the most costly to synthesize amino acid, to the initiation of chromosome replication [13].
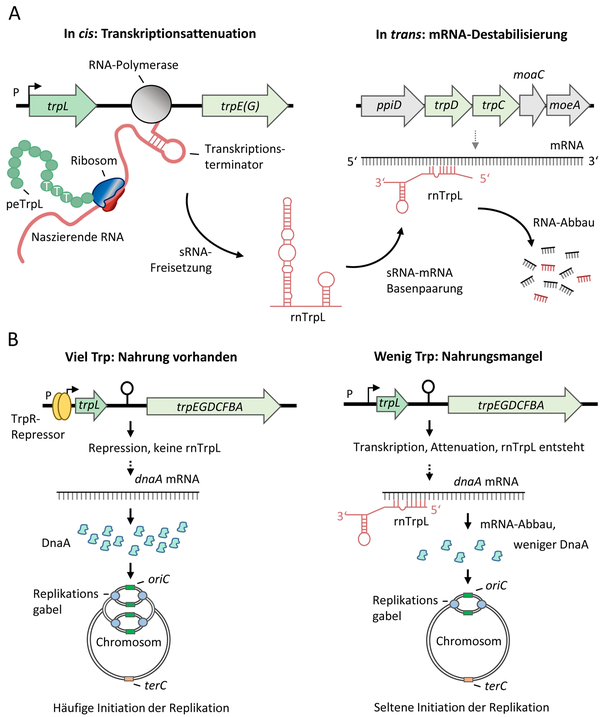
Trp-related roles of the attenuator sRNA rnTrpL in S. meliloti (A) and E. coli (B). From: Melior H and Evguenieva-Hackenberg E (2021) RNA-Regulation und Antibiotikaresistenz: Trans-agierende Attenuator-RNA und Leaderpeptid in Bakterien. Biospektrum, 02.21:127-130, DOI: 10.1007/s12268-021-1545-0
Role of the trp attenuator and its trans-acting products in adaptation to antibiotic exposure.
The attenuator sRNA rnTrpL is produced not only under conditions of tryptophan shortage, but also upon translation inhibition. Upon exposure to subinhibitory tetracycline (Tc) amount, transcription of trpE(G) is attenuated even under conditions of Trp insufficiency [14]. Our work suggests that the specificity of the sRNA rnTrpL is reprogrammed in the presence of Tc and that peTrpL has an own role in regulation of an antibiotic resistance operon [14-16].
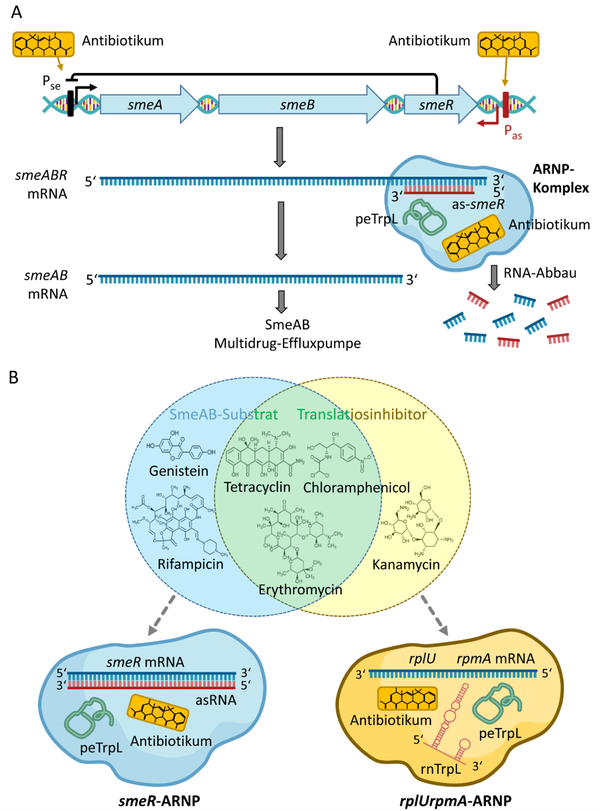
Antibiotic-related roles of peTrpL and rnTrpL in rhizobia. A) posttranscriptional regulation of the smeABR operon by peTrpL and an antisense RNA. B) Two types of peTrpL-containing ARNP complexes, one of them involved in reprograming of the sRNA rnTrpL. From: Melior H and Evguenieva-Hackenberg E (2021) RNA-Regulation und Antibiotikaresistenz: Trans-agierende Attenuator-RNA und Leaderpeptid in Bakterien. Biospektrum, 02.21:127-130, DOI: 10.1007/s12268-021-1545-0
References:
- Storz G, Wolf YI, Ramamurthi KS. (2014) Small proteins can no longer be ignored. Annu Rev Biochem. 83:753-77.
- Hadjeras L, Heiniger B, Maaß S, Scheuer R, Gelhausen R, Azarderakhsh S, Barth-Weber S, Backofen R, Becher D, Ahrens CH, Sharma CM, Evguenieva-Hackenberg E. (2023) Unraveling the small proteome of the plant symbiont Sinorhizobium meliloti by ribosome profiling and proteogenomics. microLife 4:uqad012.
- Čuklina J, Hahn J, Imakaev M, Omasits U, Förstner KU, Ljubimov N, Goebel M, Pessi G, Fischer HM, Ahrens CH, Gelfand MS and Evguenieva-Hackenberg E (2016) Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis - a rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genomics 17:302.
- Hahn J, Thalmann S, Migur A, von Boeselager RF, Kubatova N, Kubareva E, Schwalbe H and Evguenieva-Hackenberg E. (2017) Conserved small mRNA with an unique, extended Shine-Dalgarno sequence. RNA Biol. 14:1353-1363.
- Hahn J, Tsoy OV, Thalmann S, Čuklina J, Gelfand MS and Evguenieva-Hackenberg E. (2016) Small Open Reading Frames, Non-Coding RNAs and Repetitive Elements in Bradyrhizobium japonicum USDA 110. PLoS One 11:e0165429.
- Serganov A, Nudler E. (2013) A decade of riboswitches. Cell 152:17–24.
- Scheuer R, Dietz T, Kretz J, Hadjeras L, McIntosh M, Evguenieva-Hackenberg E. (2022) Incoherent dual regulation by a SAM-II riboswitch controlling translation at a distance. RNA Biol. 19:980-995.
- Baumgardt K, Melior H, Madhugiri R, Thalmann S, Schikora A, McIntosh M, Becker A and Evguenieva-Hackenberg E. (2017) RNase E and RNase J are needed for S-adenosylmethionine homeostasis in Sinorhizobium meliloti. Microbiology 163:570-583.
- Yanofsky (1981) Attenuation in the control of expression of bacterial operons. Nature 289:751-758.
- Melior H, Li S, Madhugiri R, Stötzel M, Azarderakhsh S, Barth-Weber S, Baumgardt K, Ziebuhr J and Evguenieva-Hackenberg E (2019) Transcription attenuation-derived small RNA rnTrpL regulates tryptophan biosynthesis gene expression in trans. Nucleic Acids Res. 47:6396–6410.
- Baumgardt K, Šmídová K, Rahn H, Lochnit G, Robledo M and Evguenieva-Hackenberg E (2016) The stress-related, rhizobial small RNA RcsR1 destabilizes the autoinducer synthase encoding mRNA sinI in Sinorhizobium meliloti. RNA Biol. 13:486-499.
- Li S, Edelmann D, Berghoff BA, Georg J and Evguenieva-Hackenberg E (2021) Bioinformatic prediction reveals posttranscriptional regulation of the chromosomal replication initiator gene dnaA by the attenuator sRNA rnTrpL in Escherichia coli. RNA Biol. 18:1324–1338.
- Melior H and Evguenieva-Hackenberg E (2021) RNA-Regulation und Antibiotikaresistenz: Trans-agierende Attenuator-RNA und Leaderpeptid in Bakterien. Biospektrum, 02.21:127-130.
- Melior H, Maaß S, Li S, Förstner KU, Azarderakhsh S, Varadarajan AR, Stötzel M, Elhossary M, Barth-Weber S, Ahrens CH, Becher D and Evguenieva-Hackenberg E (2020) The leader peptide petrpl forms antibiotic-containing ribonucleoprotein complexes for posttranscriptional regulation of multiresistance genes. mBio 11:1–22.
- Melior H, Li S, Stötzel M, Maaß S, Schütz R, Azarderakhsh S, Shevkoplias A, Barth-Weber S, Baumgardt K, Ziebuhr J, Förstner KU, Chervontseva Z, Becher D and Evguenieva-Hackenberg E (2021) Reprograming of sRNA target specificity by the leader peptide peTrpL in response to antibiotic exposure. Nucleic Acids Res. 49:2894–2915.
- Evguenieva-Hackenberg E. (2022) Riboregulation in bacteria: From general principles to novel mechanisms of the trp attenuator and its sRNA and peptide products. Wiley Interdiscip Rev RNA 13:e1696.
