RNA degradation and processing in prokaryotes
Role of RNases in the post-transcriptional gene regulation in Bacteria
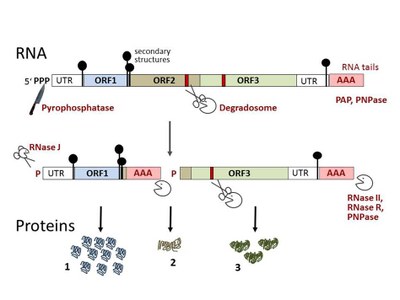
Bacteria in the environment are exposed to changing conditions and respond to the changes by flexible regulation of the gene expression. While profound but relatively slow changes in gene expression are achieved at the transcriptional level, multiple layers of posttranscriptional regulation ensure fast regulation and fine tuning. Bacterial mRNAs are usually short-lived (the mean chemical half-lives in different species are usually between 2 and 7 min), allowing fast adaptation to changing conditions.
The half-life of an mRNA is determined by the ribonucleases (RNases) in the cell and by certain RNA features which may stabilize or destabilize the molecule. An RNA molecule can be cleaved internally by endoribonucleases or can be degraded nucleotide by nucleotide from the end of the molecule by exoribonucleases, in 5’-3’ or in 3’-5’ direction. Stabilizing elements are the 5’-triphosphate of primary transcripts and strong secondary structures. RNA elements recognized by ribonucleases (RNases) like a 5’-monophosphate, specific stem-loop structures, endonucleolytic cleavage sites, and post-transcriptionally synthesized RNA tails destabilize the molecule. The short, destabilizing RNA-tails are synthesized by poly(A) polymerase or polynucleotide phosphorylase (PNPase ) in bacteria.
At the 5’-end of the RNA molecule, a monophosphate can be created by the pyrophosphatase RppH, or by internal endonucleolytic cleavages. The monophosphorylated 5’-end is efficiently recognized by key endoribonucleases like the essential RNase E in Escherichia coli. Generally, bacterial endoribonucleases cleave single-stranded AU-rich regions (RNases E/G, J and Y), or specific double stranded sites (RNase III). Endonucleolytic cleavage sites can be created, masked or de-masked by small regulatory RNAs. The resulting, destabilized fragments are cleaved further endonucleolytically, or are degraded exoribonucleolytically. Important 3’-5’ endoribonucleases with partially overlapping functions in E. coli are RNase II, RNase R and PNPase. In contrast to E. coli, exoribonucleolytic 5’-3’ degradation by RNase J takes place in Bacillus subtilis.
Often RNases are associated with protein complexes like the degradosome, which is organized by RNase E in E. coli. The major subunits of the degradosome of E. coli are RNase E, the PNPase, an RNA helicase and the enolase. The composition of the bacterial degradosome varies: In Rhodobacter capsulatus, two RNA helicases and RNase R interact with RNase E, while in B. subtilis, where no RNase E is present, the essential endoribonuclease Y organizes a degradosome containing PNPase, an RNA helicase and enolase. The degradosomes of E. coli and B. subtilis are located at the cytoplasmic membrane of the prokaryotic cell.
In our current projects we investigate the role of RNases in the Alphaproteobacteria Rhodobacter sphaeroides and Sinorhizobium meliloti. These species contain the two key bacterial RNases RNase E and RNase J, together with RNase R, RNase II, PNPase and other RNases.
Methods:
Construction and characterization of mutants in R. sphaeroides or Sinorhizobium meliloti, cloning procedures; RNA isolation; Northern blot analysis; real time RT-PCR; RNA 5’ and 3’ end detection (primer extension, RNA protection, 5’ and 3’ RACE); production, isolation and characterization of recombinant proteins; in vitro transcription, in vitro labelling of RNA, in vitro RNA degradation assays.
References
Spanka DT, Reuscher CM, Klug G. Impact of PNPase on the transcriptome of Rhodobacter sphaeroides and its cooperation with RNase III and RNase E. BMC Genomics. 2021 Feb 6;22(1):106. doi: 10.1186/s12864-021-07409-4. PMID: 33549057; PMCID: PMC7866481
Bathke J, Gauernack AS, Rupp O, Weber L, Preusser C, Lechner M, Rossbach O, Goesmann A, Evguenieva-Hackenberg E, Klug G. iCLIP analysis of RNA substrates of the archaeal exosome. BMC Genomics. 2020 Nov 16;21(1):797. doi: 10.1186/s12864-020-07200-x. PMID: 33198623 Free PMC article.
Evguenieva-Hackenberg E, Gauernack AS, Hou L, Klug G. Enzymatic Analysis of Reconstituted Archaeal Exosomes. Methods Mol Biol. 2020;2062:63-79. doi: 10.1007/978-1-4939-9822-7_4.
Spanka DT, Klug G. Maturation of UTR-Derived sRNAs Is Modulated during Adaptation to Different Growth Conditions. Int J Mol Sci. 2021 Nov 12;22(22):12260. doi: 10.3390/ijms222212260. PMID: 34830143; PMCID: PMC8625941.
Förstner KU, Reuscher CM, Haberzettl K, Weber L, Klug G. RNase E cleavage shapes the transcriptome of Rhodobacter sphaeroides and strongly impacts phototrophic growth. Life Science Alliance 2018 Aug. Vol. 1, No. 4. doi: 10.26508/lsa.201800080
Damm K, Bach S, Müller KM, Klug G, Burenina OY, Kubareva EA, Grünweller A, Hartmann RK. Improved Northern blot detection of small RNAs using EDC crosslinking and DNA/LNA probes. Methods Mol Biol. 2015;1296:41-51.
Damm K, Bach S, Müller KM, Klug G, Burenina OY, Kubareva EA, Grünweller A, Hartmann RK. Impact of RNA isolation protocols on RNA detection by Northern blotting. Methods Mol Biol. 2015;1296:29-38.
Rische-Grahl T, Weber L, Remes B, Förstner KU, Klug G. (2014) RNase J is required for processing of a small number of RNAs in Rhodobacter sphaeroides. RNA Biol. 12;11(7)
Baumgardt K, Charoenpanich P, McIntosh M, Schikora A, Stein E, Thalmann S, Kogel KH, Klug G, Becker A, Evguenieva-Hackenberg E. (2014) RNase E affects the expression of the acyl-homoserine lactone synthase gene sinI in Sinorhizobium meliloti. J Bacteriol. Apr;196(7):1435-47. doi: 10.1128/JB.01471-13. Epub 2014 Jan 31.
Rische-Grahl T, Weber L, Remes B, Förstner KU, Klug G. (2014) RNase J is required for processing of a small number of RNAs in Rhodobacter sphaeroides. RNA Biol. 12;11(7)
Baumgardt K, Charoenpanich P, McIntosh M, Schikora A, Stein E, Thalmann S, Kogel KH, Klug G, Becker A, Evguenieva-Hackenberg E. (2014) RNase E affects the expression of the acyl-homoserine lactone synthase gene sinI in Sinorhizobium meliloti. J Bacteriol. Apr;196(7):1435-47. doi: 10.1128/JB.01471-13. Epub 2014 Jan 31.
Evguenieva-Hackenberg, E., Klug, G. (2011) New aspects of RNA processing in prokaryotes. Curr. Opin. Microbiol. 14: 587-592
Evguenieva-Hackenberg, E., Klug, G. RNA degradation in Archaea and Gram-negative bacteria different from Escherichia coli. In: Molecular Biology of RNA Processing and Decay in Prokaryotes, Condon, C. ed. Prog. Mol. Biol. and Transl. Sci. 2009 85: 275-317
Purusharth RI, Klein F, Sulthana S, Jager S, Jagannadham MV, Evguenieva-Hackenberg E, Ray MK, Klug G. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005 Apr 15;280(15):14572-8.
Jager S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, Klug G. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 2001 Nov 15;29(22):4581-8.
Fritsch J, Rothfuchs R, Rauhut R, Klug G. Identification of an mRNA element promoting rate-limiting cleavage of the polycistronic puf mRNA in Rhodobacter capsulatus by an enzyme similar to RNase E. Mol Microbiol. 1995 Mar;15(6):1017-29.
Processing of ribosomal RNA (rRNA) in Alphaproteobacteria:
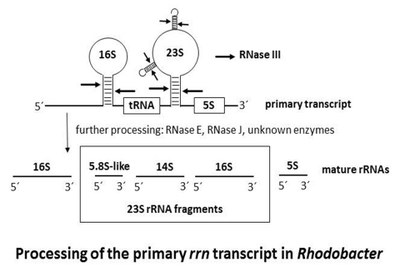
The ribosomes are the cellular machines for protein synthesis, and rRNA exhibits a central catalytic role during this process. Usually, the prokaryotic ribosomal RNAs are referred as 16S, 23S, and 5S rRNA, like in Escherichia coli. However, some bacteria have fragmented rRNA in their functional ribosomes. In many Alphaproteobacteria like Agrobacterium, Bradyrhizobium, Rhizobium, Rhodobacter and Sinorhizobium, the 23S rRNA is a precursor molecule, which is further cleaved by ribonucleases. One highly conserved processing site is near the 5’-end of the molecule. Processing at this site leads to the occurrence of a 5.8S-like rRNA in these bacteria. Cleavages at additional processing sites result in further fragmentation of the large rRNA in the 50S ribosomal subunit. In Rhodobacter capsulatus and Rhodobacter sphaeroides, the 23S rRNA precursor is cleaved into mature 5.8S-like, 14S and 16S rRNA fragments.
Investigations on the mechanisms for rRNA processing in Alphaproteobacteria revealed that the double strand-specific endoribonuclease RNase III is essential for the fragmentation of the 23S rRNA. Further processing events include RNase E and RNase J, which is responsible for the maturation of the 5’-ends of the 23S rRNA fragments. Despite this progress in our understanding of the mechanisms of rRNA fragmentation, many aspects like the maturation of the 3’-ends of rRNA and the physiological role of rRNA fragmentation are still unexplored. Recent results suggest shown that RNase J indirectly influences the maturation of 3’-ends of rRNA in R. sphaeroides.
Methods:
Construction and characterization of mutants, cloning procedures; RNA isolation; Northern blot analysis; RNA 5’ and 3’ end detection (primer extension, RNA protection, 5’ and 3’ RACE); production, isolation and characterization of recombinant proteins; in vitro transcription, in vitro labelling of RNA, in vitro RNA degradation assays.
References:
Rische, T., Klug, G. (2012) The ordered processing of intervening sequences in 23S rRNA of Rhodobacter sphaeroides requires RNase J. RNA Biology 9: 343-350
Madhugiri R, Evguenieva-Hackenberg E. RNase J is involved in the 5'-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett. 2009 Jul 21;583(14):2339-42. doi: 10.1016/j.febslet.2009.06.026. Epub 2009 Jun 21.
Evguenieva-Hackenberg E. Bacterial ribosomal RNA in pieces. Mol Microbiol. 2005 Jul;57(2):318-25. Review.
Klein F, Evguenieva-Hackenberg E. RNase E is involved in 5'-end 23S rRNA processing in alpha-Proteobacteria. Biochem Biophys Res Commun. 2002 Dec 20;299(5):780-6.
Klein F, Samorski R, Klug G, Evguenieva-Hackenberg E. Atypical processing in domain III of 23S rRNA of Rhizobium leguminosarum ATCC 10004(T) at a position homologous to an rRNA fragmentation site in protozoa. J Bacteriol. 2002 Jun;184(12):3176-85.
Conrad C, Evguenieva-Hackenberg E, Klug G. Both N-terminal catalytic and C-terminal RNA binding domain contribute to substrate specificity and cleavage site selection of RNase III. FEBS Lett. 2001 Nov 30;509(1):53-8.
Evguenieva-Hackenberg E, Klug G. RNase III processing of intervening sequences found in helix 9 of 23S rRNA in the alpha subclass of Proteobacteria. J Bacteriol. 2000 Sep;182(17):4719-29.
Conrad C, Rauhut R, Klug G. Different cleavage specificities of RNases III from Rhodobacter capsulatus and Escherichia coli. Nucleic Acids Res. 1998 Oct 1;26(19):4446-53.
Rauhut R, Jager A, Conrad C, Klug G. Identification and analysis of the rnc gene for RNase III in Rhodobacter capsulatus. Nucleic Acids Res. 1996 Apr 1;24(7):1246-51.
Selenska-Pobell S, Evguenieva-Hackenberg E. Fragmentations of the large-subunit rRNA in the family Rhizobiaceae. J Bacteriol. 1995 Dec;177(23):6993-8.
Kordes E, Jock S, Fritsch J, Bosch F, Klug G. Cloning of a gene involved in rRNA precursor processing and 23S rRNA cleavage in Rhodobacter capsulatus. J Bacteriol. 1994 Feb;176(4):1121-7.
The RNA-degrading exosome in the hyperthermophilic archaeon Sulfolobus solfataricus
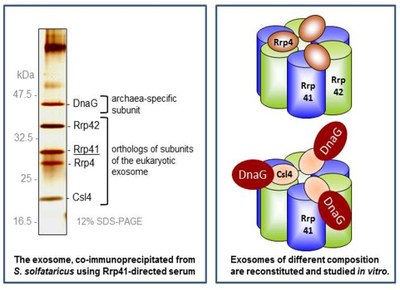
The archaeal exosome is the major exoribonuclease and the polynucleotidylation enzyme in the hyperthermophilic and acidophilic archaeon Sulfolobus solfataricus. It is a protein complex, which degrades RNA phosphorolytically in 3’ to 5’ direction. In a reverse reaction, it synthesizes heteropolymeric RNA tails using nucleoside diphosphates.
The structure of the archaeal exosome shows similarity to the structure of the bacterial polynucleotide phosphorylase (PNPase) and the eukaryotic exosome. The catalytic core of the archaeal exosome is formed by alternating Rrp41 and Rrp42 polypeptides, arranged in a hexameric ring. A flexible RNA binding cap composed of the evolutionarily conserved proteins Rrp4 and/or Csl4 is bound at the top of the ring. This cap is involved in recruitment of substrates and in interactions with other proteins. The protein complex contains an archaea-specific subunit annotated as DnaG, which directly interacts with Csl4. Together with Rrp4, DnaG confers poly(A)-specificity to the exosome. Like the bacterial degradosome, the archaeal exosome shows a subcellular localization: It is mainly localized at the periphery of the cell.
Currently we are analysing the physiological role of the archaeal exosome, the mechanisms of subcellular localization and the role of its individual subunits for interaction with different substrates and other proteins.
Methods:
in vitro transcription, in vitro labelling of RNA, in vitro RNA degradation and polyadenylation assays, Northern blot analysis, cloning, work with recombinant proteins and protein complexes, cultivation of S. solfataricus, purification and analysis of native protein complexes, Western blot analysis, co-immunoprecipitation.
References:
Gauernack AS, Lassek C, Hou L, Dzieciolowski J, Evguenieva-Hackenberg E, Klug G. Nop5 interacts with the archaeal RNA exosome. FEBS 2017 Dec;591(24):4039-4048. doi: 10.1002/1873-3468.12915. Epub 2017 Nov 28.
Hou, L., Klug, G., Evguenieva-Hackenberg, E. (2013) The archaeal DnaG protein needs Csl4 for binding to the exosome and enhances its interaction with adenine-rich RNA. RNA Biology 10: 415-424
Evguenieva-Hackenberg E, Hou L, Glaeser S, Klug G. (2014) Structure and function of the archaeal exosome. Wiley Interdiscip Rev RNA. 2014 Apr 30. doi: 10.1002/wrna.1234.
Hou, L., Klug, G., Evguenieva-Hackenberg, E. (2013) The archaeal DnaG protein needs Csl4 for binding to the exosome and enhances its interaction with adenine-rich RNA. RNA Biology 10: 415-424
Evguenieva-Hackenberg E, Bläsi U. Attack from both ends: mRNA degradation in the crenarchaeon Sulfolobus solfataricus. Biochem Soc Trans. 2013 Feb 1;41(1):379-83. doi: 10.1042/BST20120282.
Witharana, C., Roppelt, V., Lochnit, G., Klug, G., Evguenieva-Hackenberg E. (2012) Heterogeneous complexes of the RNA exosome in Sulfolobus solfataricus. Biochimie 94: 1578-1587.
Evguenieva-Hackenberg, E., Roppelt, V., Lassek, C., Klug, G. (2011) Subcellular localisation of RNA degrading proteins and protein complexes in prokaryotes. RNA Biology 8: 49-54
Roppelt, V., Klug, G., Evguenieva-Hackenberg, E. The evolutionarily conserved subunits Rrp4 and Csl4 confer different substrate specificities to the archaeal exosome. FEBS Lett. 2010 584: 2931-2936
Roppelt, V., Hobel, C.F., Albers, S.V., Lassek, C., Schwarz, H., Klug, G., Evguenieva-Hackenberg, E. The archaeal exosome localizes to the membrane. FEBS Lett. 2010 584:2791-5
Evguenieva-Hackenberg E, Roppelt V, Finsterseifer P, Klug G. Rrp4 and Csl4 are needed for efficient degradation but not for polyadenylation of synthetic and natural RNA by the archaeal exosome. Biochemistry. 2008 16;47(50):13158-68.
Walter P, Klein F, Lorentzen E, Ilchmann A, Klug G, Evguenieva-Hackenberg E. Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2006 Nov;62(4):1076-89.
Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus. EMBO Rep. 2005 Dec;6(12):1188-93.
Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005 Jul;12(7):575-81.
Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003 Sep;4(9):889-93.
Gauernack AS, Lassek C, Hou L, Dzieciolowski J, Evguenieva-Hackenberg E, Klug G. Nop5 interacts with the archaeal RNA exosome. FEBS 2017 Dec;591(24):4039-4048. doi: 10.1002/1873-3468.12915. Epub 2017 Nov 28.
Rische, T., Klug, G. (2012) The ordered processing of intervening sequences in 23S rRNA of Rhodobacter sphaeroides requires RNase J. RNA Biology 9: 343-350
Madhugiri R, Evguenieva-Hackenberg E. RNase J is involved in the 5'-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett. 2009 Jul 21;583(14):2339-42. doi: 10.1016/j.febslet.2009.06.026. Epub 2009 Jun 21.
Evguenieva-Hackenberg E. Bacterial ribosomal RNA in pieces. Mol Microbiol. 2005 Jul;57(2):318-25. Review.
Klein F, Evguenieva-Hackenberg E. RNase E is involved in 5'-end 23S rRNA processing in alpha-Proteobacteria. Biochem Biophys Res Commun. 2002 Dec 20;299(5):780-6.
