Forschungsgebiet
Forschung Allgemein
English information below
Unser zentrales Forschungsgebiet liegt in den zellulären und molekularen Mechanismen der Entwicklung von Lymphozyten, vor allem T-Zellen, im physiologischen und pathophysiologischen Kontext.
Die Bildung von T-Zellen ist ein dynamischer entwicklungsbiologischer Prozess von erheblicher klinischer Relevanz. Er hängt ab von der Migration von Knochenmark-Vorläuferzellen in den Thymus. Dort erfolgt die Spezifizierung der T-Zell-Linie, die Bildung des T-Zell-Antigen-Rezeptor-Repertoires und die Selektion von T-Zellen, die nicht-funktionelle sowie autoreaktive Zellen eliminiert. Phasen der Rekombination und Selektion sind unterbrochen durch Phasen massiver Proliferation. Aufgrund fortschreitender Involution des Thymus nimmt die Neubildung von T-Zellen mit zunehmendem Alter ab. Diese Abnahme ist ein erhebliches Problem im Rahmen einer hämatopoetischen Stammzelltransplantation. Konditionierungstherapien zerstören das Immunsystem des Patienten, während gleichzeitig die Neubildung von T-Zellen massiv verzögert abläuft. Eine ausgedehnte Phase der Immundefizienz ist die Folge. Die Verbesserung der T-Zell-Regeneration bzw. die „Verjüngung“ des alternden Thymus ist daher von fundamentaler Bedeutung für die Reduktion unerwünschter Wirkungen der Stammzelltransplantation. Darüber hinaus führt eine Fehlregulation des T-Zell-Entwicklungsprogramms häufig zur Ausbildung von Leukämien.
In diesem Kontext verfolgen wir im Wesentlichen drei Arbeitsschwerpunkte:
1) Dynamik der T-Zell-Entwicklung
2) Post-transkriptionelle Regulation der Lymphozytenentwicklung
3) Experimentelle Modelle für humane T-Zell-Entwicklung
Our Research
In order to maintain an efficient line of defense against pathogens, cells of the immune system are continually replenished throughout our life. Rapid regeneration of the immune system after hematopoietic stem cell transplantation (HSCT) is also a major prerequisite to prevent serious complications such as the emergence of fatal viral, bacterial, and fungal infections.
Generation of fresh immune cells is a highly dynamic process and prone to errors, which might ultimately result in development of cancers, such as leukemias, immunodeficiency or autoimmune disease, such as type 1 diabetes, multiple sclerosis, or rheumatoid arthritis. Thus, in depth understanding of the underlying molecular and cellular mechanisms underlying immune cell development are paramount for devising improved therapies for a broad range of immune-system related diseases. Furthermore, development of immune cells undergoes profound changes with increasing age coinciding with declining overall functionality making understanding immune cell development an important goal for medicine in an aging society.
Our laboratory is primarily interested in understanding molecular and cellular mechanisms underlying the formation of T cells. T cells are at the center of the adaptive immune response and direct the immune system to adequately respond to each of the near unlimited variety of pathogens. They are also critical for the formation of immunological memory and, thus, a key target of vaccine development. In addition to these so-called conventional T cells, other “unconventional” T cells exist with a broad range of functions. A certain subset of T cells, so-called regulatory T cells (Treg), help to prevent autoaggression of the immune system. Innate-like T cells, such as invariant Natural Killer T cells (iNKTs) or Mucosa-associated Invariant T cells (MAIT) straddle the realms of innate and adaptive immunity. Little is known about their functions in the host defense against pathogens or immunoregulatory and homeostatic functions.
Specifically, we are working on three aspects of T-cell development.
1) Lymphocyte developmental dynamics
2) Post-transcriptional control of lymphocyte development
3) Models for human T-cell development
Lymphocyte developmental dynamics
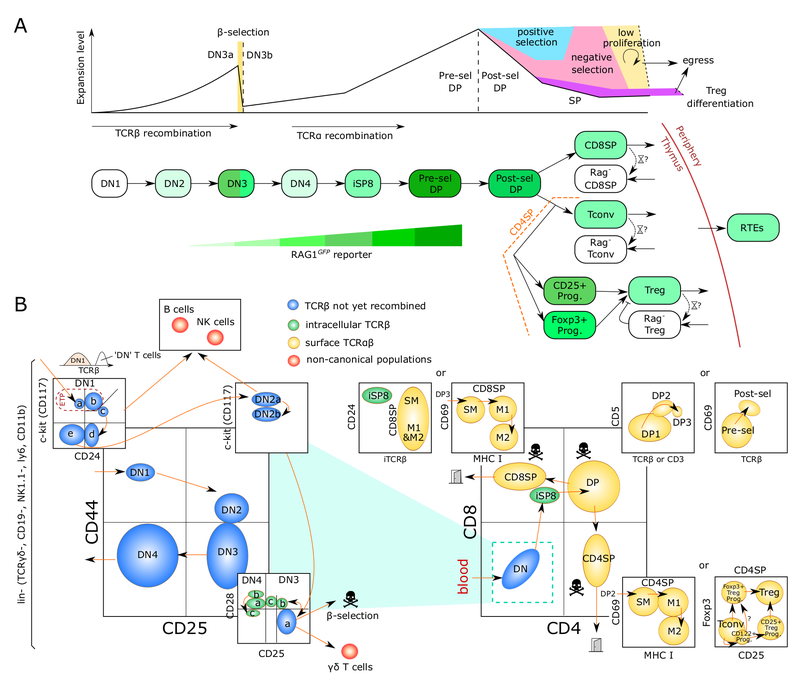
T cells, unlike other blood cells, finalize development in the thymus. We are trying to understand migration of progenitor cells from bone marrow to the thymus in molecular and quantitative terms. To this end, we have recently identified two receptors, CC chemokine receptors 7 and 9, which coordinate colonization of the thymus by progenitor cells. In addition, we have characterized these progenitors by defining minimal common characteristics. We demonstrated that indeed multiple different population act as physiologic thymus seeding progenitors. Recently, we employed a novel approach of multi-congenic fate mapping to quantitate the number of intrathymic niches for thymus seeding progenitors and defined feedback mechanisms that control thymus colonization. Currently, we are developing models of tissue homeostasis in thymus at high resolution. Our experiments have implications for designing pre-transplant conditioning regimens efficiently supporting thymus colonization. Ultimately our experiments are aimed at establishing a complete quantitative spatio-temporal model of early T cell development.
Key publications:
Coppin E, Sundarasetty BS, Rahmig S, Blume, Verheyden NA, Bahlmann F, Ravens S, Schubert U, Schmid J, Ludwig S, Geissler K, Guntinas-Lichius O, von Kaisenberg C, Groten T, Platz A, Naumann R, Ludwig B, Prinz I, Waskow C*, Krueger A* (2021). Enhanced differentiation of functional human T cells in NSGW41 mice with tissue-specific expression of human interleukin-7. Leukemia, 35(12):356. * Equal contribution
Robert PA, Kunze-Schumacher H, Greiff V, Krueger A (2021). Modeling the Dynamics of T-Cell Development in the Thymus. Entropy 23(4):437.
Krueger A, Ziętara N, Łyszkiewicz M (2017). T cell development by the numbers. Trends Immunol 38:128-139.
Ziętara N, Łyszkiewicz M, Puchałka J, Witzlau K, Förster R, Pabst O, Prinz I, Krueger A (2015). Multicongenic fate mapping quantification of dynamics of thymus colonization. J Exp Med 212:1589-1601.
Łyszkiewicz M, Ziętara N, Föhse L, Puchałka J, Diestelhorst J, Witzlau J, Prinz I, Schambach A, Krueger A (2015). Limited niche availability suppresses murine intrathymic dendritic-cell development from non-committed progenitors. Blood 125:457-64.
Post-transcriptional control of lymphocyte development
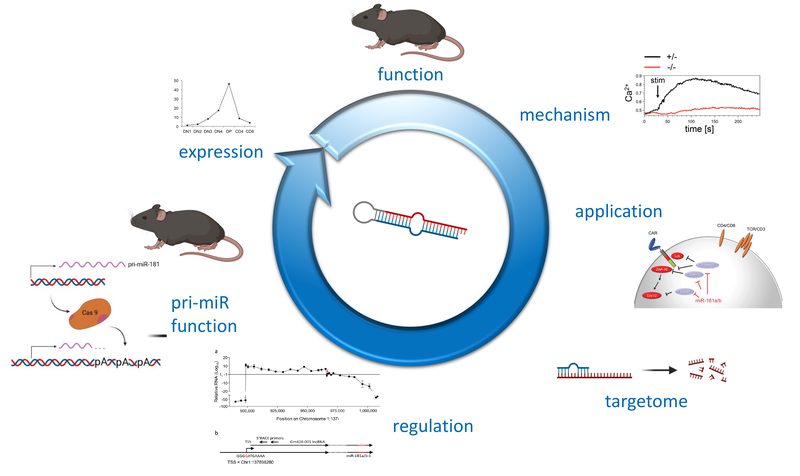
Non-coding (nc)RNAs, such as microRNAs have emerged as a novel layer of post-transcriptional gene regulation. Our laboratory is interest how a progenitor cell’s decision to become a T cell or another lymphoid or non-lymphoid cell type is controlled by ncRNA. To this end, we have conducted microRNA gene expression screens at lineage decision checkpoints and we have begun to establish regulatory networks comprising both, microRNAs and transcriptional regulators. In order to assess the function of microRNAs identified in these screens in vivo we employ classical gene targeting as well as CRISPR/Cas9-mediated deletion. We have shown that deletion of microRNA miR-181a/b-1 causes a profound defect in the generation of unconventional T cells. Furthermore, we have recently demonstrated that a cluster of microRNAs, miR-17~92 also termed (oncomiR-1 because of its function as an oncogene), is critical for early T-cell development. Ultimately, our experiments aim at integrating post-transcriptional gene regulation in a broader gene regulatory context. Understanding post-transcriptional gene regulatory programs will help to better understand molecular mechanisms of tumor formation and may result in the identification of novel therapeutic targets.
Key publications:
Krueger A, Łyszkiewicz M, Heissmeyer V (2022). Post-transcriptional control of T-cell development in the thymus. Immunol Lett 247:1-12.
Grewers Z, Krueger A (2020). MicroRNA miR-181 - A Rheostat for TCR Signaling in Thymic Selection and Peripheral T-Cell Function. Int J Mol Sci. 21(17):6200.
Łyszkiewicz M, Winter SJ, Witzlau K, Föhse L, Brownlie R, Puchałka J, Verheyden NA, Kunze-Schumacher H, Imelmann E, Blume J, Raha S, Sekiya T, Yoshimura A, Frueh JT, Ullrich E, Huehn J, Weiss S, Gutierrez MG, Prinz I, Zamoyska R, Ziętara N*, Krueger A* (2019). miR-181a/b-1 controls thymic selection of Treg cells and tunes their suppressive capacity. PLoS Biol 17(3):e2006716. * Equal contribution.
Winter SJ, Kunze-Schumacher H, Imelmann I, Grewers Z, Osthues T, Krueger A (2019). MicroRNA miR-181a/b-1 controls MAIT cell development. Immunol Cell Biol. 97(2):190-202.
Ziętara N, Łyszkiewicz M, Witzlau K, Naumann R, Hurwitz R, Langemeier J, Bohne J, Sandrock I, Ballmaier M, Weiss S, Prinz I*, and Krueger A* (2013). Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc. Natl. Acad. Sci. USA 110:7407-7412. * Equal contribution.
Models for human T-cell development
Mice with a human immune system constitute excellent models to further our understanding of the development and function of the human immune system, especially with regard to human pathology. However, current models need to be improved to overcome cross-species barriers. To this end, we have generated a BAC-transgenic mouse expressing human IL-7 under control of endogenous gene regulatory elements. Crossed onto the NSGW41 background, this model permits more efficient T-cell development with physiological ratios of B and T cells in the periphery and a diverse TCR repertoire. Strikingly, this model, termed NSGW41hIL7, permitted regeneration of mucosal immunity. This mouse model is well suited for studying human immune regeneration as well as infectious diseases of human specific pathogens and therefore provides an excellent platform for a broad range of potential collaborations.
Key publication:
Coppin E, Sundarasetty BS, Rahmig S, Blume, Verheyden NA, Bahlmann F, Ravens S, Schubert U, Schmid J, Ludwig S, Geissler K, Guntinas-Lichius O, von Kaisenberg C, Groten T, Platz A, Naumann R, Ludwig B, Prinz I, Waskow C*, Krueger A* (2021). Enhanced differentiation of functional human T cells in NSGW41 mice with tissue-specific expression of human interleukin-7. Leukemia, 35(12):356. * Equal contribution
