Research Details, Friedhoff
- Molecular enzymology of the DNA mismatch repair (MMR) pathway
-
Molecular enzymology of the DNA mismatch repair (MMR) pathway
MMR is a critical DNA repair system guarding the genome against mutation that arise from errors of the replication machinery in conjunction to important roles in DNA damage signalling and recombination. The basic machinery is highly conserved from bacteria to man. In humans, errors in the machinery lead to a predisposition to a familial (colon) cancer, known as Lynch syndrome. MMR cascade of ATPases that read out the mismatch, and ensure the repair of the newly synthesized strand specifically. In E. coli the process is initiated when the MutS protein recognizes a mismatch or looped out base. MutS then releases the mismatch and forms a stable ATP-dependent sliding clamp that is able to active MutL, the next ATPase in the cascade. This activated state of MutL then is required to activate the nuclease activity of MutH that cleaves the new strand only, followed by the loading of a helicase, UvrD. In mammals, there are close homologs to MutS and MutL, but rather than homodimers, these are heterodimeric and the nuclease activity is intrinsic to the MutL homologs, rather than a separate protein.
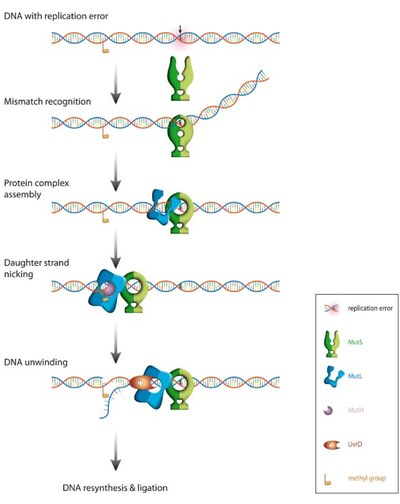
Correction of replication errors in the bacterial model system Escherichia coli:
Repair of mismatch require a sequential series of steps starting with mismatch recognition by MutS, ATP-dependent formation of a ternary complex between DNA, MutS and MutL, strand discrimination via activation of the latent endonuclease MutH, unwinding of the erroneous strand by the helicase UvrD. The unwound single-stranded DNA is degraded and replaced with a newly synthesized DNA. Note that the distance between the mismatch and the strand discrimination signal can be up to 1000 bps.The basic steps in MMR are reasonably understood, but how these molecular machines activate each other and how they translate on DNA between the mismatch and the point that signals the new strand (a hemimethylated GATC in E. coli) remains highly controversial. Noteworthy, even the temporal composition and stoichiometry of the relevant active complexes is still under debate.
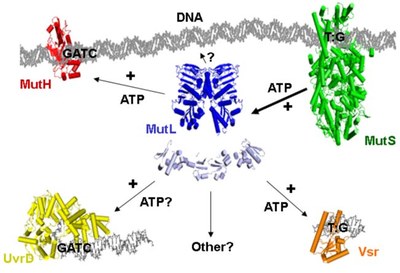
Structures of the MMR-components
Although the structures of individual components have been determined between 1998 and 2006, we still do not know the structure of complexes formed during the multistep process of MMR. We aim to determine these structure using a combination of chemical, biochemical, biophysical and computational methods.
Understanding of the basic steps in this machinery will help to pinpoint the roles of these proteins in MMR, but also promote understanding of their more elusive roles in signalling and recombination. Importantly, MMR is a potential drug target in selected synthetic lethal drug combinations that target cancer cells similar to the successful combination of PARP and BRCA1.
- Publications AG Friedhoff
-
Selected Publications of the last 5 years
1. Perevozchikova, S.A., Trikin, R.M., Heinze, R.J., Romanova, E.A., Oretskaya, T.S., Friedhoff, P. and Kubareva, E.A. (2014) Is thymidine glycol containing DNA a substrate of E. coli DNA mismatch repair system? PLoS One, 9, e104963.
2. Groothuizen, F.S., Fish, A., Petoukhov, M.V., Reumer, A., Manelyte, L., Winterwerp, H.H., Marinus, M.G., Lebbink, J.H., Svergun, D.I., Friedhoff, P. et al. (2013) Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation. Nucleic Acids Res., 41, 8166-8181.
3. Cristovao, M., Sisamakis, E., Hingorani, M.M., Marx, A.D., Jung, C.P., Rothwell, P.J., Seidel, C.A. and Friedhoff, P. (2012) Single-molecule multiparameter fluorescence spectroscopy reveals directional MutS binding to mismatched bases in DNA. Nucleic Acids Res., 40, 5448-5464.
4. Winkler, I., Marx, A.D., Lariviere, D., Heinze, R.J., Cristovao, M., Reumer, A., Curth, U., Sixma, T.K. and Friedhoff, P. (2011) Chemical trapping of the dynamic MutS-MutL complex formed in DNA mismatch repair in Escherichia coli. J. Biol. Chem., 286, 17326-17337.
5. Pillon, M.C., Lorenowicz, J.J., Uckelmann, M., Klocko, A.D., Mitchell, R.R., Chung, Y.S., Modrich, P., Walker, G.C., Simmons, L.A., Friedhoff, P. et al. (2010) Structure of the endonuclease domain of MutL: unlicensed to cut. Mol. Cell, 39, 145-151.
- Collaboration partners
-
Collaboration partners (in alphabetic order)
Janusz Bujnicki, International Institute of Molecular and Cell Biology in Warsaw, Poland
Ute Curth, Mediziniche Hochschule Hannover, Germany
Dagmar Klostermeier, Westfälische Wilhelms-Universtät, Münster, Germany
Elena Kubareva – Lomonossov State University, Moscow, Russia
Damien Lariviere – Fondation Fourmentin-Guilbert, Noisy le grand, France
Joyce Lebbink – Erasmus Medical School, Rotterdam, The Nederlands
Titia Sixma – Netherlands Cancer Institute (NKI), Amsterdam, The Nederlands
Terence Strick – Institut Jacques Monod (CNRS), Paris, France
