Role of non-coding RNAs in beneficial symbioses
Ena Šečić
Research focus:
ncRNAomics to uncover beneficial dialogues in plant symbioses - Ena Šečić
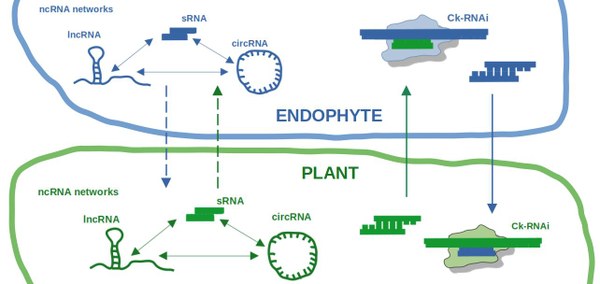
Background: Non-coding (nc) RNAs are important regulators of plant development and stress responses. In the interaction with microbes plant ncRNAs are induced to regulate endogenous gene expression. In addition, many classes of ncRNAs are also expressed in eukaryotic microbes as part of their development and in the process of plant colonization. Additionally, the specific class of small (s) ncRNAs were shown to be transferred from plant to microbe and vice versa, in order to regulate gene expression in a cross-kingdom manner. Defined as cross-kingdom (ckRNAi), this regulation involves transport and subsequent utilization of the target organism RNAi machinery to silence target mRNAs. The vast majority of plan-microbe systems in which such ncRNA regulation is studied are plant-pathogen interactions. In these systems, ncRNAs of the pathogen regulate immunity-related genes of the host plant, while the host plant ncRNAs modulate virulence or essential genes of the pathogen. Thus, ncRNAs are versatile players in cellular homeostasis, as well as agile regulators of gene expression during biotic interactions.
Project: Our aim is to study ncRNA-based regulation of gene expression during establishment and maintenance of beneficial plant symbioses with root endophytes. For that purpose, we use the model endophyte Serendipita indica (syn. Piriformospora indica), which can be cultivated axenically and has a wide host range. This allows for detection of ncRNA classes across a multitude of plant hosts, including model plants and staple crops within monocot and dicot clade. In order to understand the broad landscape of ncRNA networks induced and possibly exchanged between plants and endophytes, we focus on sRNAs, long non-coding (lnc)RNAs and circular (circ)RNAs, as well as mRNAs that these ncRNA networks regulate. To make the impact of our findings applicable in durable plant protection strategies, we conduct specific screens for ncRNAs with beneficial effects on host plants, and facilitate the translation of this knowledge from model to staple crop plants by comparative analyses of plant hosts colonized by Serendipita indica.
Lab tools / techniques: ckRNAi-seq, degradome-seq, mRNA-seq, comparative epigenomics, ncRNA assays, AGO pull-downs.