Research Details, Sträßer
- Coupling of single steps in gene expression
-
Coupling of single steps in gene expression
Gene Expression is a fundamental process of any living cell that brings the genetic information stored in the DNA to life. In eukaryotic cells, the genetic information is transcribed into messenger (m)RNA by RNA polymerase II (RNAPII). Already co-transcriptionally, the mRNA is processed (capped, spliced, and polyadenylated). In addition, proteins bind to the mRNA and package into a mature mRNP (see Figure 1). Because nucleoplasm and cytoplasm are separated by the nuclear envelope in eukaryotic cells, the mRNP needs to be exported through the nuclear pore complexes (NPCs) to the cytoplasm (see Figure 1). Here, the information encoded in the mRNA is translated into a polypeptide chain by the ribosomes. Eventually, the mRNA is degraded (see Figure1).
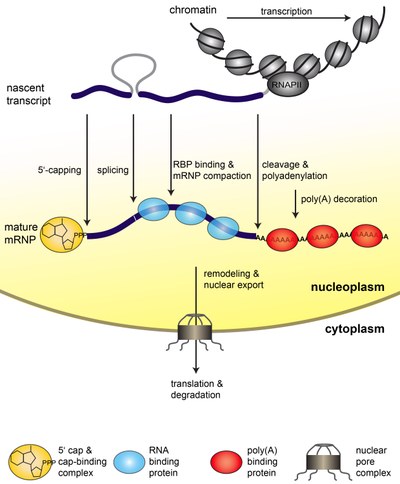
Figure 1. Steps of nuclear mRNP formation. The mRNA is synthesized from the protein coding gene by RNAPII as a nascent transcript. The nascent transcript is processed while transcription elongation still takes place: It is capped, spliced, cleaved and polyadenylated. In addition, mRNA-binding proteins bind to the mRNA and package it into an mRNP. These mRNA-binding proteins influence mRNA stability and are necessary for nuclear export. After a likely remodeling of the mRNP it is exported to the cytoplasm through the nuclear pore complex (NPC). In the cytoplasm the mRNA is translated by the ribosomes, which synthesize the encoded protein. Eventually, the mRNA is degraded.
The single steps of mRNP formation do not occur consecutively, but are tightly interconnected. This coupling guarantees an efficient and accurate expression of the genetic information and thus provides a quality control mechanism for this fundamental cellular process. In recent years, multiple links between the intranuclear steps of gene expression have been shown. One of the key players in this coupling is the so called TREX complex, which couples transcription and mRNA export. It consists of the heteropentameric THO complex (consisting of the components Tho2, Hpr1, Mft1, Thp2, and Tex1), the nuclear mRNA export factors Sub2 and Yra1, and the SR-proteins Gbp2 and Hrb1. TREX travels along the transcribed gene with RNA polymerase II, binds the nascent mRNA, and “feeds” it into the mRNA export pathway via its subunits and mRNA export factors Sub2 and Yra1 (see Figure 2). Yra1 then directly binds the mRNA exporter Mex67-Mtr2. Mex67-Mtr2 in turn binds directly to the mRNA as well as to nuclear pore proteins and transports the mRNP through the nuclear pore complex to the cytoplasm (see Figure 2).
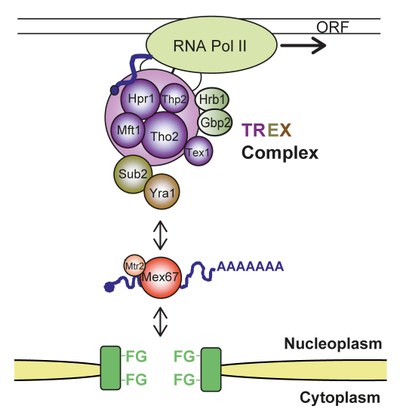
Figure 2. The TREX complex couples transcription to nuclear export of the mRNP. TREX consists of the subcomplex THO (a heteropentameric complex, consisting of the proteins Tho2, Hpr1, Mft1, Thp2, and Tex1), the SR-proteins Gbp2 and Hrb1, and the intranuclear mRNA export factors Sub2 and Yra1. Yra1 interacts directly with the mRNA exporter Mex67-Mtr2, which binds to the mRNA and the proteins of the nuclear pore complex (NPC). Mex67-Mtr2 transports the mRNP through the NPC to the cytoplasm.
We propose that the single steps of nuclear mRNP formation are organized within a molecular mRNP packaging station (see Figure 3). This molecular packaging station might be built of the C-terminal domain of Rpb1, the largest subunit of RNAPII (Rpb1-CTD), the transcription elongation factor Spt5 with its C-terminal region (CTR), the mRNA and TREX (see Figure 3). The CTD consists of heptapeptide repeats with the sequence YSPTSPS, and its differential phosphorylation – mainly on S5 and S2 – is known to recruit different factors needed for mRNP formation. The mRNP packaging station coordinates the single steps of nuclear mRNP formation providing efficiency and quality control to this process.
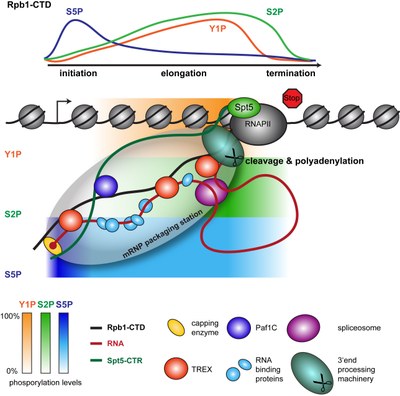
Figure 3. The mRNP packaging station. As the mRNA emerges from RNAPII during transcription elongation it is capped, spliced, cleaved and polyadenylated (processing). In addition, a multitude of RNA-binding proteins binds to the mRNA, packaging it in an mRNP. We hypothesize that these single steps of mRNP formation are coordinated and controlled within an mRNP packaging station. This mRNP packaging station is assembled by three recruitment platforms and possibly additional coordinating proteins such as the TREX complex, which ensure spatial proximity of all processes. The three recruitment platforms for mRNA processing and mRNA-binding proteins are indicated by different colors (red: RNA, black: Rpb1-CTD and green: Spt5-CTR). The different mRNA processing and mRNA-binding proteins are defined in the legend below the figure. The capping enzyme is exchanged for the cap binding complex consisting of Cbc20 and Cbc80 after capping is complete. The differential phosphorylation of the CTD (C-terminal domain of Rpb1, the largest subunit of RNAPII) is indicated at the top as well as below the chromatin (blue: S5P, orange: Y1P, green: S2P). The phosphorylation status is indicated by a gradient from 0-100% as indicated by the bars in the bottom left hand corner. 0% reflects no detectable phosphorylation while 100% reflects the maximum signal observed at an average gene in a ChIP experiment.
Our long term goal is to understand how gene expression is regulated by the cross communication between its single steps especially during mRNP formation. We would like to understand, how an mRNP is formed, how it is remodelled, how other concomitant steps (transcription elongation, the chromatin state, mRNA processing) influence the composition of the mRNP, and how its composition effects downstream processes such as translation and mRNA degradation.
Ongoing subprojects include:
1. Nuclear mRNP formation and remodelling
2. A novel factor in RNAPIII transcription
3. Analysis of gene expression on a systemic level
Figure 4. Cartoon of the mRNP packaging station.
- Selected Publications of the last 5 years
-
Selected Publications of the last 5 years
Karakasili, E.*, Burkert-Kautzsch, C.*, Kieser, A., and Sträßer, K. (2014) Degradation of DNA damage-independently stalled RNA polymerase II is independent of the E3 ligase Elc1. Nucleic Acids Res. 42: 10503-10515
* equal contribution
Meinel, D.M., Burkert-Kautzsch, C., Kieser, A., O’Duibhir, E., Siebert, M., Mayer, A., Cramer, P., Söding, J., Holstege, F.C., and Sträßer, K. (2013) Recruitment of TREX to the Transcription Machinery by its Direct Binding to the Phospho-CTD of RNA Polymerase II. PLoS Genetics 9: e1003914
Chanarat, S. and Sträßer, K. (2013) Splicing and beyond: The many faces of the Prp19 complex. BBA-MCR 1833: 2126-2134 (review)
Chanarat, S., Seizl, M., and Sträßer, K. (2011) The Prp19 Complex is a Novel Transcription Elongation Factor Required for TREX Occupancy at Transcribed Genes. Genes Dev. 25: 1147-1158
Lammens, K.*, Bemeleit, D.J.*, Möckel, C.*, Clausing, E., Schele, A., Hartung, S., Schiller, C.B., Lucas, M., Angermüller, C., Söding, J., Sträßer, K., and Hopfner, K.-P. (2011) The Mre11:Rad50 complex shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145: 54-66
* equal contribution
Clausing, E., Mayer, A.*, Chanarat, S.*, Müller, B., Germann, S.M., Cramer, P., Lisby, M., and Sträßer, K. (2010) The transcription elongation factor Bur1-Bur2 interacts with Replication Protein A to maintain genome stability. J. Biol. Chem. 285: 41665-41674
* equal contribution
Röther, S.*, Burkert, C.*, Brünger, K.M.*, Mayer, A.*, Kieser, A., and Sträßer, K. (2010) Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA 16: 1393-1401
* equal contribution
- Collaboration partners (alphabetic order)
-
Collaboration partners (alphabetic order)
Ruedi Aebersold, Swiss Federal Institute of Technology (ETH), Zurich, Switzerland
Traude Beilharz, Monash University, Melbourne, Australia
Hong Cheng, Shanghai Institutes for Biological Sciences, Shanghai, China
Sofia Georgieva, Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russia
Riccardo Pellarin, Institut Pasteur, Paris, France
Henning Urlaub, Max Planck Institute of Biophysical Chemistry, Göttingen
