B07
Selective biomarkers for right heart hypertrophy and failure
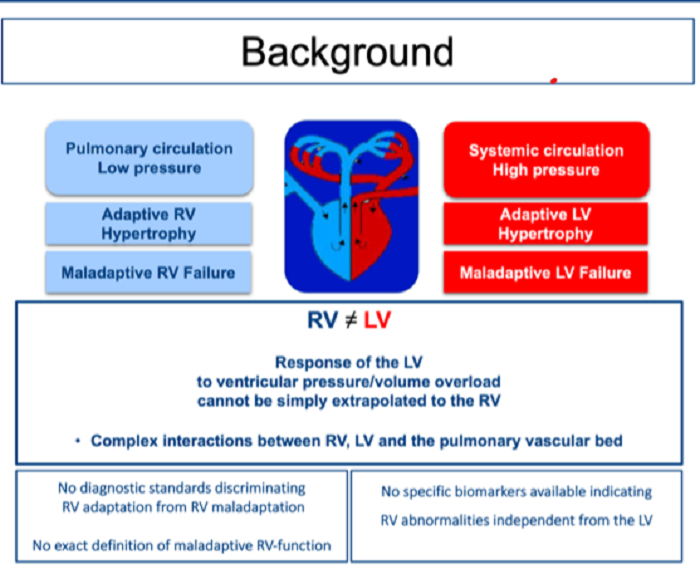
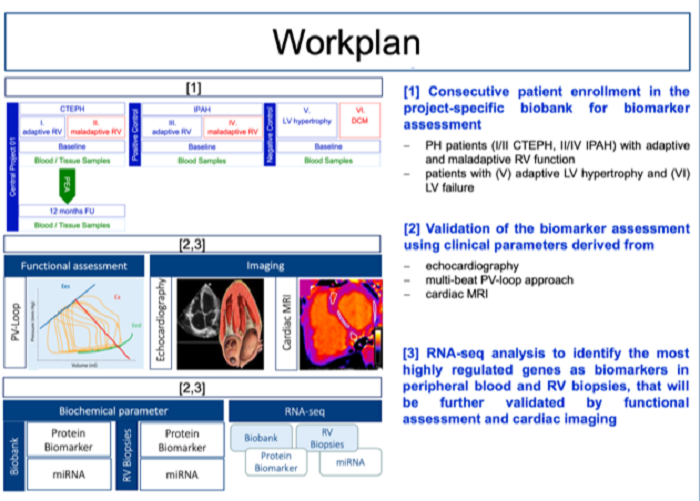
We recently identified promising biomarkers that discriminate adaptive right ventricular (RV) hypertrophy from maladaptive RV failure in human pulmonary hypertension. In addition, results from RNA-sequencing analysis of human RV biopsies identified highly regulated genes that also correlate with clinical parameters in the investigated patients. In a next step, our recent results will be further related to the established clinical, imaging, and functional parameters in PH patients that are of diagnostic and prognostic interest. In addition, we aim to further investigate cellular transformation processes that lead to fibrotic alterations in different animal models.