Regulation of repetitive sequences and genome maintenance
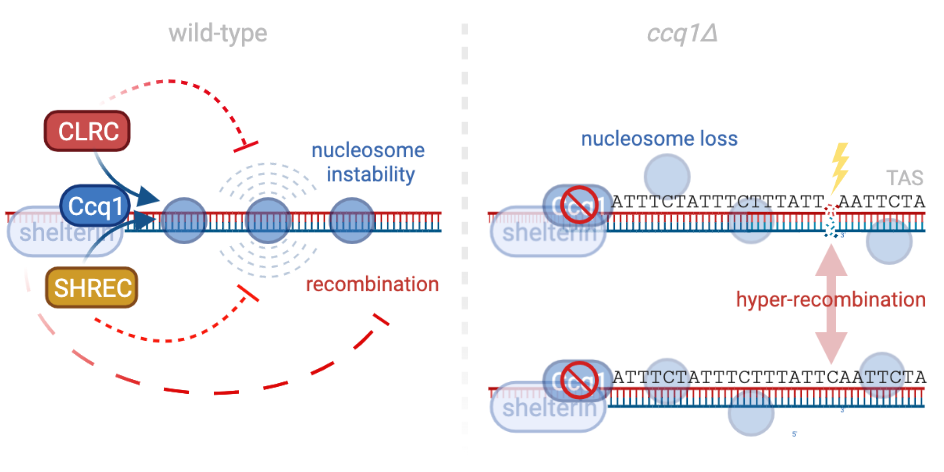
DNA repeats are often embedded into heterochromatin to prevent unwarranted recombination and genome instability. However, during DNA damage or loss, recombination of these repeats may become necessary. We showed recently that repeat-like sequences present at subtelomeres display metastable nucleosomes and are hyper-recombinogenic, promoting telomere plasticity and recombination (van Emden, Forn et al., 2019). This is an intrinsic property of the underlying DNA sequences and counteracted by Ccq1, a member of the conserved shelterin complex protecting the chromosome ends (telomeres). We are currently testing whether these fragile DNA sequences play an active role in telomere maintenance independent of telomerase.
DNA repair and recombination of other repetitive genomic sequences also need to be properly controlled. The ribosomal RNA encoding genes are organized in repetitive units (rRNA repeats) and tethered to the inner nuclear membrane by a protein complex known as CLIP. This prevents their release from the nucleolus and their recombination by the DNA repair machinery, which is excluded from the nucleolus. However, their release becomes necessary when DNA damage within the rRNA repeats occur. We discovered, in Saccharomyces cerevisiae (baker’s or budding yeast), that upon DNA damage the CLIP complex becomes first phosphorylated, followed by the addition of the small-ubiquitin-like modifier SUMO. SUMOylation of CLIP serves as recognition signal for the conserved AAA ATPase Cdc48 (p97 in mammals), which untether the rDNA repeats from CLIP, resulting in their release and repair outside the nucleolus. Remarkably, as we demonstrated, this mechanism of rRNA release is also conserved in human cells (Capella et al., Nature comms 2021).
